1. Introduction
Infectious diseases remain a major global health problem, which cause many deaths from illnesses like tuberculosis, malaria and HIV/AIDS. These diseases spread quickly, especially in areas with poor healthcare systems, making the situation worse. Current diagnostic methods often don't work well because they are too slow and not sensitive enough. This leads to treatment delays and helps diseases be spreading.
PCR technology changed how we diagnose diseases when it was developed in the 1980s. This method makes copies of DNA very quickly, allowing doctors to find germs with great accuracy. It works even when there are very few germs present, which helps doctors diagnose infections early and choose the right treatments.
New versions of PCR work even better. Digital PCR can count exact amounts of DNA, while quantitative PCR lets scientists watch the copying process as it happens. The newest methods combine PCR with CRISPR technology, like the PCR-Cas13a test that finds Helicobacter pylori very well. Other new tools, like special microscopes, help scientists see these tiny biological processes.
This review looks at how PCR has changed from a research tool to an important medical test. It shows PCR's good points, like being very sensitive and accurate, but also its problems, like needing expensive machines and sometimes giving wrong results. New developments include smaller portable machines, tests that find multiple germs at once, and using computers to help read results.
2. Digital PCR in pathogen detection: advances and challenges
Infectious diseases remain a major worldwide health problem. Dangerous germs cause illnesses like HIV/AIDS, tuberculosis, and malaria. Together, these diseases infect more than 100 million people each year. Doctors need accurate tests to identify these infections. Good tests must correctly tell which germ is making someone sick. Without proper testing, treatment can be delayed or wrong. Current diagnostic methods try to solve this problem, so they look for specific parts of each germ. This helps doctors choose the right medicine faster. Conventional diagnostic approaches exhibit three primary limitations: reduced sensitivity in low pathogen load scenarios (typically <100 CFU/mL), prolonged processing times (often 48-72 hours), and insufficient specificity leading to false-positive interpretations.
PCR changed how people find microbes because it is very sensitive. It can detect 10-100 copies of pathogen DNA in a sample. This is 100 to 1000 times better than older culture methods. qPCR is the most common type used today, which measures DNA as it copies in real-time. However, it only gives relative amounts using Ct values, not exact numbers. The method works by copying specific DNA pieces many times. This makes even small amounts of pathogen DNA easier to find. Doctors and scientists use it because it's reliable and fast.
Digital PCR (dPCR) platforms address this quantification limitation through absolute target measurement. By partitioning samples into thousands of discrete reactions, dPCR achieves precise nucleic acid counting without requiring standard curves. Clinical studies show PCR can detect single DNA copies reliably. This high sensitivity helps doctors in two important ways. Firstly, it allows monitoring how patients respond to treatment for long-term infections. Secondly, it identifies new antibiotic resistance early. The method works well for chronic diseases like HIV and hepatitis. It also spots when bacteria stop responding to drugs. These uses make PCR valuable for patient care and public health. Hospitals now use this technology routinely, because it gives accurate results when older tests might miss infections, which helps doctors make better treatment decisions [1].
3. The application of PCR
Infectious diseases remain a major worldwide health problem. Dangerous germs cause illnesses like HIV/AIDS, tuberculosis, and malaria. Together, these diseases infect more than 100 million people each year. Doctors need accurate tests to identify these infections. Good tests must correctly tell which germ is making someone sick. Without proper testing, treatment can be delayed or wrong. Current diagnostic methods try to solve this problem, so they look for specific parts of each germ. This helps doctors choose the right medicine faster. Conventional diagnostic approaches exhibit three primary limitations: reduced sensitivity in low pathogen load scenarios (typically <100 CFU/mL), prolonged processing times (often 48-72 hours), and insufficient specificity leading to false-positive interpretations.
PCR changed how people find microbes because it is very sensitive. It can detect 10-100 copies of pathogen DNA in a sample. This is 100 to 1000 times better than older culture methods. qPCR is the most common type used today, which measures DNA as it copies in real-time. However, it only gives relative amounts using Ct values, not exact numbers. The method works by copying specific DNA pieces many times. This makes even small amounts of pathogen DNA easier to find. Doctors and scientists use it because it's reliable and fast.
Digital PCR (dPCR) platforms address this quantification limitation through absolute target measurement. By partitioning samples into thousands of discrete reactions, dPCR achieves precise nucleic acid counting without requiring standard curves. Clinical studies show PCR can detect single DNA copies reliably. This high sensitivity helps doctors in two important ways. Firstly, it allows monitoring how patients respond to treatment for long-term infections. Secondly, it identifies new antibiotic resistance early. The method works well for chronic diseases like HIV and hepatitis. It also spots when bacteria stop responding to drugs. These uses make PCR valuable for patient care and public health. Hospitals now use this technology routinely, because It gives accurate results when older tests might miss infections, which helps doctors make better treatment decisions [2].
Quantitative PCR (qPCR) is widely used in clinical labs to measure virus levels. It counts viral DNA accurately from 10 to 100 million copies per milliliter. This helps doctors monitor infections and treatments. In forensic labs, PCR analyzes DNA fingerprints. It looks at short repeating DNA sections. The test is so precise that the chance of a wrong match is less than one in a billion. Environmental scientists use PCR to study bacteria. By copying the 16S rRNA gene, they can identify 97-99% of bacterium types correctly, which helps understand soil and water ecosystems.
The Cancer researchers also rely on PCR. It finds rare cancer mutations in mixed cell samples. The test spots mutations present in just 1 out of 1000 cells. This precision helps guide cancer treatment decisions.
Modern PCR systems have overcome early limitations through three key innovations. First, device miniaturization now allows point-of-care testing in non-laboratory settings. Second, multiplexing capabilities enable concurrent detection of 10-20 pathogen targets in a single reaction. Third, CRISPR system integration has improved specificity through sequence-specific recognition. These technological improvements have reduced reagent and equipment costs by 40% compared to 2010 prices. Sensitivity remains uncompromised, with detection limits maintained at 1-10 target copies per reaction. Ongoing research prioritizes two areas: establishing uniform testing protocols across platforms and adapting systems for detection of emerging viral and bacterial pathogens [3].
Quantitative PCR (Q-PCR) measures target DNA concentration by detecting fluorescence signals during amplification cycles. This process occurs in real-time, allowing quantification during the exponential phase when amplicon production correlates directly with initial template quantity. Exponential phase analysis prevents inaccuracies found in end-point PCR, where final product amounts often misrepresent original target abundances due to PCR amplification biases.
Fluorescence-based Q-PCR demonstrates superior performance compared to earlier methods. It achieves higher sensitivity and broader dynamic range, reliably detecting twofold concentration differences. Previously employed techniques like competitive PCR and MPN-PCR required extensive post-amplification processing and showed greater variability. The current fluorescence detection system eliminates these additional steps while providing more accurate quantification results, particularly for mixed-template samples where amplification efficiency varies between targets [4]. Figure 1 shows another kinds of fluorescence-based PCR: single-molecule fluorescence resonance energy transfer PCR (smFRET PCR). The experimental procedure involved immobilizing Cy3-labeled target DNA on a surface, then observing its interaction with Cy5-labeled guide RNA. Results showed the engineered Cas9 variants require more precise base-pair matching for DNA binding, enhancing their specificity while maintaining functionality. Additionally, the figure presents Jeon et al.'s smFRET/TIRFM study revealing Cas12a's dynamic binding and cutting behavior at 0.13-second intervals. These single-molecule observations provide valuable insights into CRISPR systems' target recognition mechanisms at nanometer resolution.
PCR technology has become essential for identifying animal sources in dairy products. This method works better than protein tests because DNA stays stable during processing. Scientists use different PCR types to check milk ingredients. Multiplex PCR can find several animal types at once. Real-time PCR with TaqMan probes gives exact numbers of DNA copies. New digital PCR methods make measurements even more precise.
These tests help find fake or mixed milk products. They work for common milk like cow and goat, and special types like camel and yak. The methods are fast and cost less than older techniques. Recent improvements use special primers that work for many animals. This makes testing easier and more reliable for food inspectors. The tests now handle complex milk products like cheese and yogurt well [5].
PCR technology requires carefully designed primers for successful DNA amplification. Primer design involves multiple critical parameters. The optimal length ranges between 18-22 base pairs. The melting temperature should be 55-65°C. The GC content needs to stay within 40-60%. Secondary structures must be avoided.
Early solutions included commercial software packages like OLIGO and PrimerSelect. Later, open-source alternatives became available. These tools automate complex calculations. They significantly improve design success rates. Computer-designed primers achieve 90-95% success in first attempts. Manual designs typically show only 60-70% success. The automated approach saves 50-70% of optimization time. It maintains high specificity standards required for research and diagnostics.
Modern primer design software handles all necessary calculations. It evaluates potential binding sites. It checks for mismatches. It predicts secondary structures. This automation reduces human error. It increases experimental reproducibility. These tools have become essential for molecular biology workflows
PCR technology has greatly improved parasite studies in the last 15 years. It can detect single parasite cells, making it very sensitive. Researchers now use PCR for three main areas: classifying parasites, studying how diseases spread, and examining parasite-host relationships. Two PCR methods are especially useful. First, multiplex PCR uses several primer sets together. This allows detection of different parasites in one sample. Second, real-time PCR measures exactly how many parasites are present. Both methods help scientists diagnose and track parasitic diseases more effectively [6].
Real-time PCR gives faster results than regular PCR. It uses special probes like TaqMan or dyes like SYBR Green to measure DNA as it copies. This method needs only 2-3 hours, while old methods took 6-8 hours. It also gives exact numbers of DNA copies, from just 1-5 up to millions. The results are very consistent, with less than 5% variation between tests. These improvements help doctors and researchers. They get answers faster and know exactly how much DNA is present. The tests work for many purposes, like finding infections or studying genes. The equipment does most of the work automatically, making fewer mistakes than manual methods [7].
PCR is widely used to identify bacteria in clinical labs. There are two main types. First, real-time PCR finds specific bacteria fast, usually in 2-3 hours. It works well for known bacteria and resistance genes. Second, 16S rRNA sequencing finds unknown bacteria. It looks at a common gene part present in all bacteria. The 16S method takes longer but finds more types of bacteria. It can detect as few as 10-100 bacterial cells per sample. New fast PCR methods make 16S tests quicker. Old tests needed 1-3 minutes per step. New tests need only 25 seconds per step. This makes machines available sooner for other tests [8].
Helicobacter pylori infection remains a significant clinical challenge, with conventional diagnostic methods exhibiting notable limitations. Culture-based detection, while specific (90-95%), demonstrates variable sensitivity (70-90%) and requires 3-7 days for completion. Urea breath tests offer non-invasive alternatives but show reduced accuracy (85-95%) in patients with recent proton pump inhibitor use. The recently developed PCR-Cas13a assay addresses these limitations through a two-stage detection process: initial isothermal amplification of target DNA followed by CRISPR-Cas13a mediated sequence recognition. Clinical validations report a detection threshold of 1 copy/μL with a 30-minute turnaround time, representing a 6-12-fold improvement over conventional PCR. However, implementation barriers persist, including equipment requirements and the unresolved issue of antibiotic resistance observed in 20-30% of clinical isolates.
In parallel, mechanical biosensing platforms have achieved pN-scale resolution through nanoelectromechanical system (NEMS) innovations. Single-molecule force spectroscopy now enables simultaneous measurement and manipulation of individual biomolecules. Atomic force microscopy continues to provide essential structural insights, with high-speed variants (HS-AFM) achieving 10-50 fps imaging through miniaturized cantilevers and optimized feedback systems. These advances facilitate real-time observation of dynamic processes including DNA-protein interactions and molecular motor activity at sub-nanometer resolution [9].
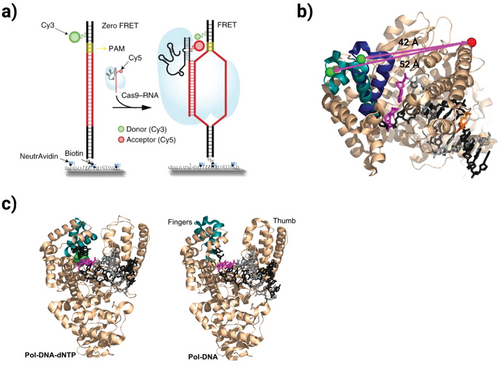
Figure 1: This figure demonstrates the use of single-molecule FRET (smFRET) technology to compare the DNA-targeting specificity between engineered Cas9 variants (eCas9 and Cas9-HF1) and wild-type Cas9 [9]
4. Advancements and future prospects of PCR technology
The Polymerase Chain Reaction (PCR) has revolutionized molecular diagnostics and biomedical research, with its future trajectory pointing toward enhanced precision, automation, and multifunctionality. Emerging PCR variants, including quantitative real-time PCR (qPCR) and droplet digital PCR (ddPCR), offer unparalleled sensitivity in nucleic acid quantification, enabling early detection of pathogens, oncogenic mutations, and genetic disorders.
PCR technology requires carefully designed primers for successful DNA amplification. Primer design involves multiple critical parameters. Early solutions included commercial software packages like OLIGO and PrimerSelect. Later, open-source alternatives became available. These tools automate complex calculations. They significantly improve design success rates. Computer-designed primers achieve 90-95% success in first attempts. Manual designs typically show only 60-70% success. The automated approach saves 50-70% of optimization time. It maintains high specificity standards required for research and diagnostics.
Modern primer design software handles all necessary calculations. It evaluates potential binding sites. It checks for mismatches. It predicts secondary structures. This automation reduces human error. It increases experimental reproducibility. These tools have become essential for molecular biology workflows [10].
Despite its advantages, PCR technology has some constraints. The accuracy of results can be affected by poor sample quality or contamination, potentially leading to false readings. Additionally, certain substances in test samples may inhibit the PCR reaction, requiring careful experimental design. Future improvements may address these issues through automated systems that minimize human error and enhanced detection methods that distinguish between similar genetic sequences. Researchers are also developing portable PCR devices for field use in remote areas, which could revolutionize disease surveillance. As these innovations progress, PCR will likely maintain its central role in biological research while expanding into new areas like personalized medicine and environmental monitoring. The ongoing refinement of this technology promises to further transform scientific discovery and healthcare delivery in the coming decade.
A key development lies in miniaturized, point-of-care PCR systems, which integrate microfluidics and isothermal amplification to facilitate rapid, field-deployable diagnostics—critical for infectious disease surveillance in resource-limited settings. Furthermore, multiplex PCR assays are evolving to simultaneously detect numerous targets with high specificity, improving efficiency in clinical and epidemiological screening.
5. Conclusion
PCR was developed in the 1980s and changed how we detect pathogens. Old diagnostic methods often missed infections or took too long. New PCR methods work better. There are two main types: qPCR and dPCR. qPCR measures DNA as it copies. dPCR is more precise. A special kind called ddPCR splits samples into tiny drops. It counts DNA pieces one by one. This helps find very small amounts of pathogens. It also studies genetic differences well. These methods give fast, accurate results. They find infections old tests missed. Doctors can diagnose patients quicker. The tests work for many diseases. They are now used in hospitals and labs worldwide.
New PCR methods now work with other technologies to do more. The PCR-Cas13a test uses CRISPR to find pathogens like H. pylori very accurately. At the same time, special microscopes called AFM help scientists see tiny biological structures. The newest AFMs take pictures very fast, showing how molecules move and interact.
PCR tests are used in many areas. Doctors use them to diagnose diseases. Police use them to solve crimes. Scientists use them to check water and soil. New improvements let PCR tests find many targets at once. The CRISPR system helps make the tests more precise. These changes will help doctors give better treatments and track disease outbreaks.
References
[1]. Lei S, Chen S, Zhong Q. Digital PCR for accurate quantification of pathogens: Principles, applications, challenges and future prospects. Int J Biol Macromol. 2021 Aug 1;184:750-759. doi: 10.1016/j.ijbiomac.2021.06.132. Epub 2021 Jun 24. PMID: 34171259.
[2]. Wang Y, Liu L, Liu X, Wu K, Zhu X, Ma L, Su J. An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori. J Pers Med. 2022 Dec 17;12(12):2082. doi: 10.3390/jpm12122082. PMID: 36556302; PMCID: PMC9784247.
[3]. Chu J, Romero A, Taulbee J, Aran K. Development of Single Molecule Techniques for Sensing and Manipulation of CRISPR and Polymerase Enzymes. Small. 2023 Sep;19(38):e2300328. doi: 10.1002/smll.202300328. Epub 2023 May 24. PMID: 37226388; PMCID: PMC10524706.
[4]. Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. 2009 Jan;67(1):6-20. doi: 10.1111/j.1574-6941.2008.00629.x. PMID: 19120456.
[5]. Ma X, Xia H, Pan Y, Huang Y, Xu T, Guan F. Double-Tube Multiplex TaqMan Real-Time PCR for the Detection of Eight Animal-Derived Dairy Ingredients. J Agric Food Chem. 2024 May 22;72(20):11640-11651. doi: 10.1021/acs.jafc.4c01294. Epub 2024 May 9. PMID: 38725129; PMCID: PMC11117397.
[6]. Zarlenga DS, Higgins J. PCR as a diagnostic and quantitative technique in veterinary parasitology. Vet Parasitol. 2001 Nov 22;101(3-4):215-30. doi: 10.1016/s0304-4017(01)00568-4. PMID: 11707298.
[7]. Pedlar M, Emery MJ, Warburton PJ. Amplifying PCR productivity and environmental sustainability through shortened cycling protocols. Biochimie. 2024 Jun;221:60-64. doi: 10.1016/j.biochi.2024.01.013. Epub 2024 Jan 21. PMID: 38262587.
[8]. Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004 Jun;4(6):337-48. doi: 10.1016/S1473-3099(04)01044-8. PMID: 15172342; PMCID: PMC7106425.
[9]. Galluzzi L, Ceccarelli M, Diotallevi A, Menotta M, Magnani M. Real-time PCR applications for diagnosis of leishmaniasis. Parasit Vectors. 2018 May 2;11(1):273. doi: 10.1186/s13071-018-2859-8. PMID: 29716641; PMCID: PMC5930967.Guo J, Starr D,
[10]. Guo H. Classification and review of free PCR primer design software. Bioinformatics. 2021 Apr 1;36(22-23):5263-5268. doi: 10.1093/bioinformatics/btaa910. PMID: 33104196.
Cite this article
Li,Z. (2025). The Principle, Application and Development Prospect of the Technology PCR. Theoretical and Natural Science,112,35-41.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICEGEE 2025 Symposium: Sensor Technology and Multimodal Data Analysis
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Lei S, Chen S, Zhong Q. Digital PCR for accurate quantification of pathogens: Principles, applications, challenges and future prospects. Int J Biol Macromol. 2021 Aug 1;184:750-759. doi: 10.1016/j.ijbiomac.2021.06.132. Epub 2021 Jun 24. PMID: 34171259.
[2]. Wang Y, Liu L, Liu X, Wu K, Zhu X, Ma L, Su J. An Ultrasensitive PCR-Based CRISPR-Cas13a Method for the Detection of Helicobacter pylori. J Pers Med. 2022 Dec 17;12(12):2082. doi: 10.3390/jpm12122082. PMID: 36556302; PMCID: PMC9784247.
[3]. Chu J, Romero A, Taulbee J, Aran K. Development of Single Molecule Techniques for Sensing and Manipulation of CRISPR and Polymerase Enzymes. Small. 2023 Sep;19(38):e2300328. doi: 10.1002/smll.202300328. Epub 2023 May 24. PMID: 37226388; PMCID: PMC10524706.
[4]. Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. 2009 Jan;67(1):6-20. doi: 10.1111/j.1574-6941.2008.00629.x. PMID: 19120456.
[5]. Ma X, Xia H, Pan Y, Huang Y, Xu T, Guan F. Double-Tube Multiplex TaqMan Real-Time PCR for the Detection of Eight Animal-Derived Dairy Ingredients. J Agric Food Chem. 2024 May 22;72(20):11640-11651. doi: 10.1021/acs.jafc.4c01294. Epub 2024 May 9. PMID: 38725129; PMCID: PMC11117397.
[6]. Zarlenga DS, Higgins J. PCR as a diagnostic and quantitative technique in veterinary parasitology. Vet Parasitol. 2001 Nov 22;101(3-4):215-30. doi: 10.1016/s0304-4017(01)00568-4. PMID: 11707298.
[7]. Pedlar M, Emery MJ, Warburton PJ. Amplifying PCR productivity and environmental sustainability through shortened cycling protocols. Biochimie. 2024 Jun;221:60-64. doi: 10.1016/j.biochi.2024.01.013. Epub 2024 Jan 21. PMID: 38262587.
[8]. Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004 Jun;4(6):337-48. doi: 10.1016/S1473-3099(04)01044-8. PMID: 15172342; PMCID: PMC7106425.
[9]. Galluzzi L, Ceccarelli M, Diotallevi A, Menotta M, Magnani M. Real-time PCR applications for diagnosis of leishmaniasis. Parasit Vectors. 2018 May 2;11(1):273. doi: 10.1186/s13071-018-2859-8. PMID: 29716641; PMCID: PMC5930967.Guo J, Starr D,
[10]. Guo H. Classification and review of free PCR primer design software. Bioinformatics. 2021 Apr 1;36(22-23):5263-5268. doi: 10.1093/bioinformatics/btaa910. PMID: 33104196.









