1. Introduction
Triple-negative breast cancer (TNBC) is cancer that lacks molecular targets such as estrogen receptors (ER) and gesterone receptors (GR). Breast cancer expressed by PR and human epidermal growth factor 2 (HER2) is one of the most common and deadly cancers in women [1]. Epidemiological data survey shows that TNBC accounts for 15%-25% of the increase in breast cancer. According to the latest global cancer burden data released by the International Agency for Research on Cancer (IARC) of the World Health Organization, the incidence of breast cancer continues to rise, increasing by 1% per year from 2012 to 2021 [2]. Compared with other breast cancer subtypes, TNBC has few treatment options, and chemotherapy is still the main treatment method for TNBC patients, and the recurrence rate is very high. Currently, only PARP inhibitors can play a role in targeted therapy, but TNBC patients with BRCA1 and BRCA2 mutations can only benefit clinically [3]. Overall, TNBC patients have a poor prognosis, short survival, very high malignancy, often middle to late diagnosis, and a high risk of visceral metastasis [4]. Therefore, it is of great clinical significance to actively search for early diagnosis and treatment of TNBC.
Several common risk factors have been identified for TNBC. These include genetic predisposition, particularly mutations in the BRCA1 and BRCA2 genes, which are associated with an increased risk of developing this subtype of breast cancer [5]. Additionally, TNBC is more prevalent in younger women, with African American and Hispanic women being disproportionately affected [6]. Lifestyle factors such as obesity, lack of physical activity, and certain reproductive history elements, including nulliparity or late age at first full-term pregnancy, have also been linked to an increased risk of TNBC [7]. Understanding these risk factors and the underlying molecular mechanisms is crucial for the development of effective prevention and treatment strategies for TNBC.
Ubiquitin is a small protein found in all eukaryotes (most eukaryotic cells) [8]. Ubiquitin is composed of 76 amino acids and has a molecular weight of about 8.451kDa [9]. Ubiquitination is a post-translational modification mediated by ubiquitin-activating enzymes, ubiquitin-ligases, and ubiquitin-conjugating enzymes, which plays a very important role in clearing misfolded or unfolded proteins and maintaining homeostasis in cells and tissues [10]. The ubiquitin-proteasome system (UPS) includes two important protein degradation pathways that maintain intracellular material balance [11]. The two systems control the cell cycle, intracellular signal transduction, DNA transcription repair, and translation, and jointly maintain the homeostasis of the intracellular environment, ultimately controlling cell quality.
RNF19A, an E3 ubiquitin ligase, has gained increasing attention due to its involvement in various cellular processes and its potential implications in cancer pathogenesis, including TNBC. RNF19A is part of the RBR (RING-between-RING) family of E3 ligases, which are responsible for transferring ubiquitin from an E2 ubiquitin-conjugating enzyme to specific substrate proteins [12]. This post-translational modification can dictate a wide array of outcomes for the substrate, including degradation via the proteasome, alteration of cellular location, or modulation of activity. Chenglei found that NLRP11 attenuates Toll-like receptor signaling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A [13]. Recent studies have highlighted RNF19A's role in regulating protein homeostasis, cell cycle progression, and stress responses. RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination [14]. Overexpression of RNF19A has been observed in several types of cancer, suggesting that it may contribute to tumorigenesis by promoting the degradation of tumor suppressor proteins or the stabilization of oncogenic factors [15]. Specifically, in the context of breast cancer, RNF19A is thought to influence pathways associated with cell survival and proliferation, although its exact role in TNBC remains to be fully elucidated. Moreover, the identification of RNF19A's substrates has been a critical area of research, as understanding these interactions could reveal novel aspects of its function and regulation. Investigating changes in the subcellular localization of these substrates via immunofluorescence staining can provide insights into their biological roles and how alterations in these roles may contribute to cancer development and progression [16]. As research progresses, RNF19A may emerge as a potential therapeutic target, offering new avenues for intervention in TNBC and other malignancies.
This study’s aim is to investigate the subcellular localization changes of potential substrates of the E3 ubiquitin ligase RNF19A using immunofluorescence staining. Given the critical role of RNF19A in protein ubiquitination and its emerging significance in cancer biology, understanding how its substrates are spatially distributed within the cell provides important insights into their functional regulation [17]. Changes in substrate localization may reflect post-translational modifications mediated by RNF19A, as well as its involvement in regulating key cellular pathways. Furthermore, this research provides a platform for identifying novel substrates of RNF19A and evaluating their roles in processes such as proliferation, apoptosis, and metastasis. The findings will not only expand the understanding of RNF19A's functional complexity but also help explore its potential as a therapeutic target for TNBC and other diseases.
2. Method
2.1. Cell culture
For A549, a human lung adenocarcinoma epithelial cell line, a medium that supplied the necessary nutrients and growth factors to maintain its epithelial characteristics was used. In contrast, for MDA-MB-231, a human breast adenocarcinoma cell line renowned for its aggressiveness and metastatic potential, a medium that supported its rapid proliferation and phenotypic expression was opted for . The A549 and MDA-MB-231 cell lines, pivotal to this study, were meticulously cultured under strict standard conditions to preserve their physiological integrity and responsiveness. These conditions included a controlled temperature of 37°C, optimal for mammalian cell metabolism and growth, and a humidified atmosphere with a precise 5% CO₂ concentration crucial for balancing the pH within the culture medium and thus maintaining cellular homeostasis.
2.2. Experimental treatment of cells
Cells were subjected to various experimental treatments to observe the effects on the subcellular localization of RNF19A substrates. Treatments were tailored based on the study’s objectives, including the use of specific inhibitors and other compounds. SOD1 Inhibition: In an effort to dissect the influence of oxidative stress on the intracellular positioning of RNF19A substrates, cells were treated with the SOD1 inhibitor ATN-224 at a concentration of 30 μM. This intervention facilitated an assessment of how alterations in oxidative stress levels might modulate the functionality and cellular distribution of RNF19A substrates. TP53 Activation: The role of TP53, a pivotal tumor suppressor protein, was investigated by modulating its activity within the cells. Cells were treated with the TP53 activator PRIMA-1 at a concentration of 30 μM to explore its influence on RNF19A activity and the consequent localization of its substrates. This manipulation was crucial in determining the interplay between TP53 signaling and the RNF19A-mediated protein degradation pathways.
2.3. Vector construction
To construct the RNF19A-overexpressing vector, the RNF19A gene was cloned into the pCDH-Flag-c-Myc vector. Primer pairs were designed according to the guidelines provided by the ClonExpress MultiS One Step Cloning Kit (C113-01, Vazyme). The pCDH-Flag-c-Myc vector was linearized using the restriction enzymes EcoRI (ER0271, ThermoFisher Scientific) and BamHI (ER0055, ThermoFisher Scientific). The PCR-amplified RNF19A gene was then ligated with the linearized vector to form the recombinant pCDH-Flag-c-Myc-RNF19A. The recombinant plasmid was transformed into DH5α competent bacteria for amplification, followed by plasmid extraction for further use as shown in Figure 1.
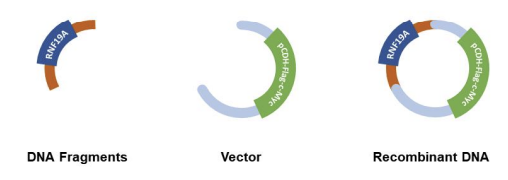
2.4. Cell transfection
For cell transfection, cells were seeded at a density of 200,000 to 700,000 per well in a six-well plate, depending on the cell type, size, and growth rate, to achieve approximately 70-90% confluence the following day. The medium was replaced with 2 ml of fresh, serum-containing medium without antibiotics. In separate sterile tubes, 125 μl of antibiotic- and serum-free DMEM or Opti-MEM® Medium (Gibco) was added. In one tube, 2.5 μg of the recombinant pCDH-Flag-c-Myc-RNF19A DNA was added and mixed gently. In the other tube, 5 μl of Lipo6000™ transfection reagent (Beyotine) was added and mixed. After a 5-minute incubation at room temperature, the DNA-containing medium was combined with the Lipo6000™ reagent medium and incubated for another 5 minutes. Then, 250 μL of the Lipo6000™-DNA mixture was added to each well and distributed evenly. The medium was replaced with fresh complete culture medium 4 hours post-transfection. Transfection efficiency was assessed by Western Blot after an additional 24 hours of culture.
2.5. Immunofluorescence staining
Immunofluorescence staining was employed to visualize the subcellular localization of RNF19A substrates. Cells were fixed, permeabilized, and incubated with primary antibodies specific to the substrates of TP53 or SOD1 antibody and anti-Flag antibody, followed by secondary antibodies conjugated to fluorescent dyes. Imaging was performed using a confocal, allowing for the detailed examination of changes in localization patterns under different experimental conditions. This method enabled the identification of potential alterations in subcellular distribution influenced by RNF19A activity.
2.6. Statistical analysis
The obtained results were subjected to statistical analysis using GraphPad Prism 6.02 software. Descriptive statistics were used to summarize the data, and the results were presented as mean ± standard deviation (SD). A one-way analysis of variance (ANOVA) was performed to assess the differences between groups. The statistical significance level was set at p < 0.05.
3. Results
3.1. Identification of RNF19A substrates
To identify the substrates of RNF19A, the Ubibrowser platform was utilized(http://ubibrowser.bio-it.cn/ubibrowser_v3). Through this investigation, six substrates were identified: RNF19A, SOD1, TP53, CASR, SNCAIP, and TRAF6 (see Table 1). In addition, other substrates of RNF19A were evaluated as illustrated in Figure 2.
Notably, SOD1 and TP53 were selected for further analysis due to their significant roles in TNBC and NSCLC, where they perform critical functions. Based on previous literatures, PRIMA-1 serves as an activator of TP53, while ATN-224 functions as an inhibitor of SOD1. Treatments using PRIMA-1 and ATN-224 were conducted, with concentration details to be specified.
|
Gene name |
Description |
Function |
Pathway |
Reference |
|
RNF19A |
E3 Ubiquitin Protein Ligase |
RNF19A help create a protein that has RING-finger motifs. |
AKT/mTOR |
[18] |
|
CASR |
Calcium Sensing receptor |
This gene encodes a Gprotein receptor that can detect changes in calcium levels in circulation |
cAMP pathway |
[19] |
|
SNCAIP |
Synuclein Alpha Interacting Protein |
This gene makes a protein have parts that allow it to interact with other proteins such as allowing a protein to bind with other proteins. |
РАК-2p34 |
[20] |
|
SOD1 |
Superoxide Dismutase 1 |
This gene makes a protein that can bind to copper and zinc, which allows it to destroy superoxide radicals in the body. |
Amyotrophic lateral sclerosis (ALS) |
[21] |
|
TP53 |
Cellular tumor antigen p53 |
This gene makes a protein that is capable of supressing tumour as it can regulate expression of genes. |
ATM-CHK2-p53 |
[22] |
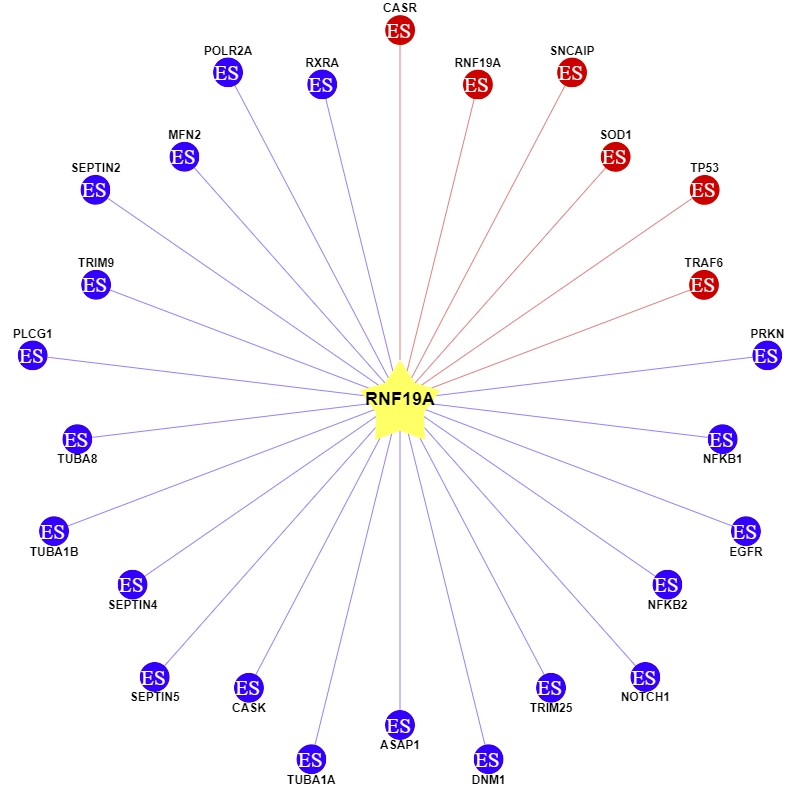
3.2. Establishment of cell lines for substrate validation
To verify the potential function of substrates of RNF19A, two types of cell expression lines were established: Vector Control (VC) and RNF19A Overexpression (RNF19A). The expression of RNF19A in these cell lines was confirmed by Western blot analysis.
As shown in Figure 3, a distinct band at approximately 90 kDa was detected in the A549 and MDA-MB-231 cell lines, indicating the successful expression of the RNF19A protein. In contrast, only a weak band was observed at the 90 kDa position in the vector control cells. As illustrated in the figure, RNF19A is expressed in lanes 1 and 2, whereas lanes 3 and 4 show no expression, consistent with the expectations for vector controls. This result confirms the successful overexpression of RNF19A in the A549 and MDA-MB-231 cell lines.
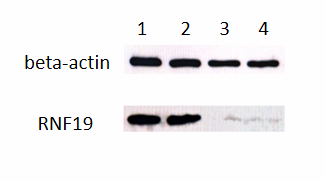
Beta-actin is always expressed.
Sample 1: A549 transfected with over expression RNF19A vector.
Sample 2: MDA-MB-231 transfected with over expression RNF19 vector.
Sample 3: A549 transfected with vector control.
Sample 4: MDA-MB-231 transfected with vector control.
3.3. Effect of TP53 activator treatment on RNF19A
Immunofluorescence microscopy was employed to observe the subcellular localization of RNF19A. The study utilized two types of constructs: Vector Control (VC) and RNF19A Overexpression (RNF19A). After transfection into A549 and MDA-MB-231 cell lines, the cells were treated with PRIMA-1 (30 μM).
Using the confocal, its clear that the green fluorescence of RNF19A is mainly located near the blue nucleus by confocal. The overexpressed cell lines are compared to the control cell lines, indicating that the green fluorescence signal was reduced in RNF19A-overexpressing cells following treatment with PRIMA-1 compared to untreated cells as shown in Figure 4A.
Quantitative analysis of relative fluorescence intensity for RNF19A in Figure 4B revealed a significant difference in fluorescence between vector control and overexpression conditions under basal conditions, suggesting that RNF19A alone impacts fluorescence intensity in both cell lines (A549: P < 0.01; MDA-MB-231: P < 0.01).
However, upon treatment with PRIMA-1 (30 μM), the fluorescence intensity did not change significantly in A549 cells (P < 0.01), while it decreased in MDA-MB-231 cells (P < 0.05).
Overall, the observed decrease in RNF19A fluorescence following treatment with the TP53 activator PRIMA-1 highlights the potential role of RNF19A in regulating the TP53 signaling pathway.
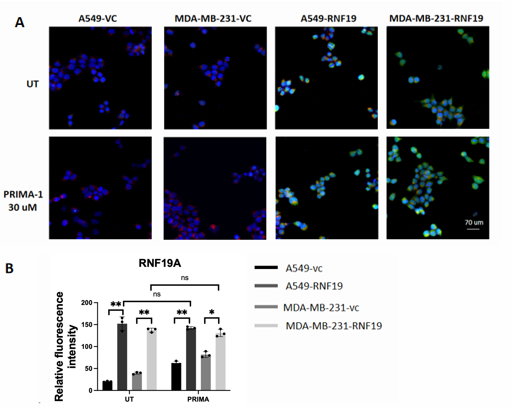
A: Untreated group and PRIMA-1 treated cell were captured under a confocal microscope.
B: Relative fluorescence intensity of RNF19A
RNF19A was stained green, TP53 was stained red, and nuclei were stained with DAPI (blue). Images were captured using confocal microscope.
*P < 0.05,**P < 0.01,***P < 0.001
3.4. Effect of SOD1 inhibitor treatment on RNF19A
To investigate the impact of a SOD1 inhibitor on RNF19A localization, the localization of RNF19A transfected with SOD1 is also seen through an immunofluorescence microscopy image. There are two plasmids involved in this study: VC and over-expressed RNF19A. The cells are treated with ATN-224 (30 μM) after transfection into A549 and MDA-MB-231 cell lines, which resulted in a clearer and sharper version of green and blue fluorescent with less orangish or red colors as seen in Figure 5A.
After analysis of the difference in Relative Fluorescence Intensity, it has been concluded that RNF19A leads to a decrease in expression for both cells. After treatment from ATN-224 (30 μM), it is obvious that there is a lowered amount of relative fluorescence intensity in A549, but the intensity of MDA-MB-231 doesn’t seem to be affected.
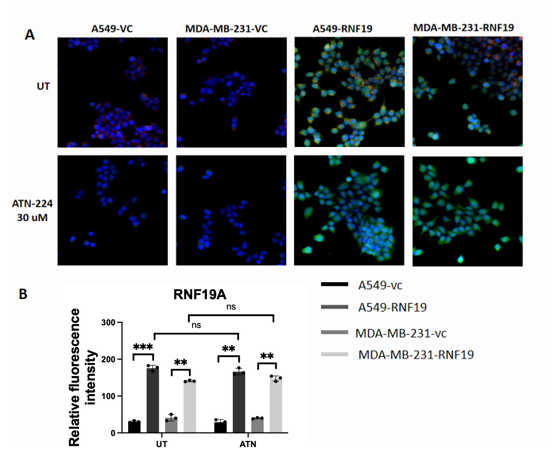
A: Untreated group and ATN-224 treated cell were captured under a confocal microscope.
B: Relative fluorescence intensity of RNF19A
RNF19A was stained green, SOD1 was stained red, and nuclei were stained with DAPI (blue). Images were captured using confocal microscope.
**P < 0.01,***P < 0.001
4. Discussion
Ubiquitination, a pivotal post-transcriptional modification, remains an underexplored target in the pharmaceutical market. Consequently, there is a pressing need for increased research efforts to elucidate the potential of drugs targeting this modification [23]. This study investigates the subcellular localization of the E3 ligase RNF19A and its potential substrates. Utilizing immunofluorescence staining as the primary technique, this research offers valuable insights into the involvement of RNF19A in the regulatory mechanisms of protein degradation and cellular pathways, revealing the regulation pattern of E3 ligase RNF19A in different types of NSCLC and TNBC as seen in Figure 6. By employing Vector Control and RNF19A Overexpression plasmids, the hypothesis was tested in A549 and MDA-MB-231 cells under TP53 activator and SOD1 inhibitor treatments. The observed alterations in RNF19A fluorescence expression underscore the pivotal role of E3 ligases in directing substrate fate and function. The variations in substrate localization patterns under different conditions suggest significant roles for these substrates in cellular processes. The results from this study indicate that RNF19A predominantly localizes near the nuclei. Other studies have identified RNF19A’s exact location in the rat’s sperm development. During spermatid development, Rnf19a and Psmc3 are initially found in Golgi-derived proacrosomal vesicles. Later on, Rnf19a, Psmc3, and ubiquitin are seen along the cytosolic side of the acrosomal membranes and the acroplaxome, a cytoskeletal plate linking the acrosome to the spermatid nuclear envelope [12]. Future investigations could explore a more exact localization of RNF19A in cancer cells.
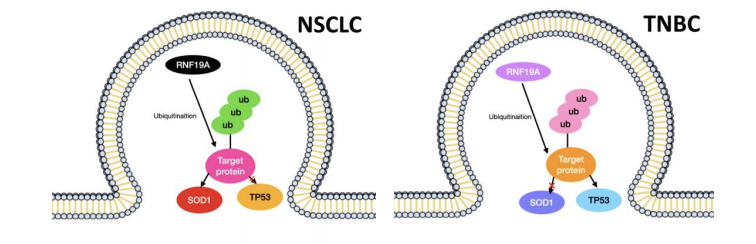
The TP53 gene, a key player in cancer development, is among the most critical tumor suppressor genes, involved in DNA repair, cell cycle regulation, and autophagy [12]. The immunostaining data suggest an interaction between RNF19A and TP53, evidencing weakly decreased RNF19A fluorescence in A549 cells, however, no significant change of expression in MDA-MB-231 cells upon TP53 activation. In Cheng Y et al.’s research, it is proven that the overexpression of RNF19A can impact the degradation of TP53 in NSCLC cells [22]. There is not clear research done on the subcellular localization of RNF19A effecting by TP53. The data supports the fact that TP53 is pretty evident in cancer cells, as it has been found in both TNBC and NSCLC.
SOD1 is traditionally associated with reactive oxygen species (ROS) activities, but recent research suggests additional functions in transcription modification and post-translational regulation [24]. Findings reveal a decrease in fluorescence intensity in A549 cells under SOD1 inhibition, with no significant change observed in MDA-MB-231, suggesting specific implications for the RNF19A-SOD1 pathway in NSCLC. Conversely, SOD1 might not significantly impact TNBC. According to previous findings, RNF19A is localized in inclusion bodies of the motor neuron cells in mutated SOD1 Amyotrophic lateral sclerosis (ALS) mouse model [25]. It has been discovered in a study done by Martin et al. that the localization of human SOD1 is found in the nuclei of a motor neuron in an ALS mouse model [26]. An interaction between RNF19A and SOD1 in breast and lung cancer cells has been found. Based on this research and other data, the subcellular localization in cancer cells and nervous cells is quite different. In this research, RNF19A’s localization after the addition of SOD1 is near the nuclear which is normal. However, in nervous cells, the localization of RNF19A after the mutation of SOD1 is abnormal. Originally found in the centrosomal region, RNF19A is instead localized in the inclusion bodies [26]. This means that the function of SOD1 is different depending on the specific type of cell, neuron, and cancer cell.
5. Conclusion
This study demonstrates that the subcellular localization of E3 ligase RNF19A and its potential substrates is highly dynamic, responding significantly to various experimental treatments. Through immunofluorescence staining, distinct localization patterns emerged, highlighting RNF19A's intricate regulatory roles in cellular processes. The findings in this study underscore the significance of RNF19A in modulating substrate function and intracellular positioning, which could be crucial for understanding its contributions to pathological conditions like cancer. Findings reveal specific implications for the RNF19A-SOD1 pathway in NSCLC. And interaction between RNF19A and TP53, further research is needed to investigate their relationship with the occurrence of breast tumors. These insights serve as a foundational basis for future research focused on exploring RNF19A as a potential therapeutic target, especially in diseases characterized by protein localization and function.
References
[1]. M.S. Moran, Radiation therapy in the locoregional treatment of triple-negative breast cancer, The Lancet. Oncology 16(3) (2015) e113-22.
[2]. A.N. Giaquinto, H. Sung, L.A. Newman, R.A. Freedman, R.A. Smith, J. Star, A. Jemal, R.L. Siegel, Breast cancer statistics 2024, CA: a cancer journal for clinicians 74(6) (2024) 477-495.
[3]. G. Bianchini, C. De Angelis, L. Licata, L. Gianni, Treatment landscape of triple-negative breast cancer - expanded options, evolving needs, Nature reviews. Clinical oncology 19(2) (2022) 91-113.
[4]. Y. Hu, C. Wang, H. Liang, J. Li, Q. Yang, The treatment landscape of triple-negative breast cancer, Medical oncology (Northwood, London, England) 41(10) (2024) 236.
[5]. A.M. Karim, J. Eun Kwon, T. Ali, J. Jang, I. Ullah, Y.G. Lee, D.W. Park, J. Park, J.W. Jeang, S.C. Kang, Triple-negative breast cancer: epidemiology, molecular mechanisms, and modern vaccine-based treatment strategies, Biochemical pharmacology 212 (2023) 115545.
[6]. S. Dawood, Triple-negative breast cancer: epidemiology and management options, Drugs 70(17) (2010) 2247-58.
[7]. I. Susumu, I. Kiyotaka, S. Shinichi, U. Keiji, H. Naoki, Y. Hiromasa, T. Yoshinori, Benefits of terminal noncardioplegic warm blood retrograde perfusion after terminal warm blood cardioplegia perfusion prior to aortic unclamping in open heart surgery, The Journal of cardiovascular surgery 47(6) (2006) 677-82.
[8]. D.H. Schlesinger, G. Goldstein, H.D. Niall, The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells, Biochemistry 14(10) (1975) 2214-8.
[9]. D.H. Schlesinger, G. Goldstein, Molecular conservation of 74 amino acid sequence of ubiquitin between cattle and man, Nature 255(5507) (1975) 423-4.
[10]. D. Popovic, D. Vucic, I. Dikic, Ubiquitination in disease pathogenesis and treatment, Nature medicine 20(11) (2014) 1242-53.
[11]. R. Kandel, J. Jung, S. Neal, Proteotoxic stress and the ubiquitin proteasome system, Seminars in cell & developmental biology 156 (2024) 107-120.
[12]. E. Rivkin, A.L. Kierszenbaum, M. Gil, L.L. Tres, Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head-tail coupling apparatus during rat spermatid development, Developmental dynamics : an official publication of the American Association of Anatomists 238(7) (2009) 1851-61.
[13]. C. Wu, Z. Su, M. Lin, J. Ou, W. Zhao, J. Cui, R.F. Wang, NLRP11 attenuates Toll-like receptor signalling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A, Nature communications 8(1) (2017) 1977.
[14]. Q. Zhu, J. Huang, H. Huang, H. Li, P. Yi, J.A. Kloeber, J. Yuan, Y. Chen, M. Deng, K. Luo, M. Gao, G. Guo, X. Tu, P. Yin, Y. Zhang, J. Su, J. Chen, Z. Lou, RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination, Nature communications 12(1) (2021) 6653.
[15]. V.U. Bai, O. Hwang, G.W. Divine, E.R. Barrack, M. Menon, G.P. Reddy, C. Hwang, Averaged differential expression for the discovery of biomarkers in the blood of patients with prostate cancer, PloS one 7(4) (2012) e34875.
[16]. K. Im, S. Mareninov, M.F.P. Diaz, W.H. Yong, An Introduction to Performing Immunofluorescence Staining, Methods in molecular biology (Clifton, N.J.) 1897 (2019) 299-311.
[17]. H. Deng, G. Ji, J. Ma, J. Cai, S. Cheng, F. Cheng, RNF19A inhibits bladder cancer progression by regulating ILK ubiquitination and inactivating the AKT/mTOR signalling pathway, Biology direct 19(1) (2024) 102.
[18]. S. Zhu, M. Tao, Y. Li, X. Wang, Z. Zhao, Y. Liu, Q. Li, Q. Li, Y. Lu, Y. Si, S. Cao, J. Ye, H3K27me3 of Rnf19a promotes neuroinflammatory response during Japanese encephalitis virus infection, Journal of neuroinflammation 20(1) (2023) 168.
[19]. H. Zuo, J. Park, A. Frangaj, J. Ye, G. Lu, J.J. Manning, W.B. Asher, Z. Lu, G.B. Hu, L. Wang, J. Mendez, E. Eng, Z. Zhang, X. Lin, R. Grassucci, W.A. Hendrickson, O.B. Clarke, J.A. Javitch, A.D. Conigrave, Q.R. Fan, Promiscuous G-protein activation by the calcium-sensing receptor, Nature 629(8011) (2024) 481-488.
[20]. Q. Zhou, M.M. Zhang, M. Liu, Z.G. Tan, Q.L. Qin, Y.G. Jiang, LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson's disease progression, Aging 13(3) (2021) 4115-4137.
[21]. R. Mohammad Malyar, H. Li, H. Enayatullah, L. Hou, R. Ahmad Farid, D. Liu, J. Akhter Bhat, J. Miao, F. Gan, K. Huang, X. Chen, Zinc-enriched probiotics enhanced growth performance, antioxidant status, immune function, gene expression, and morphological characteristics of Wistar rats raised under high ambient temperature, 3 Biotech 9(8) (2019) 291.
[22]. Y. Cheng, Y. Hu, H. Wang, Z. Zhao, X. Jiang, Y. Zhang, J. Zhang, Y. Tong, X. Qiu, Ring finger protein 19A is overexpressed in non-small cell lung cancer and mediates p53 ubiquitin-degradation to promote cancer growth, Journal of cellular and molecular medicine 25(16) (2021) 7796-7808.
[23]. P. Cohen, M. Tcherpakov, Will the ubiquitin system furnish as many drug targets as protein kinases?, Cell 143(5) (2010) 686-93.
[24]. E.C.A. Eleutherio, R.S. Silva Magalhães, A. de Araújo Brasil, J.R. Monteiro Neto, L. de Holanda Paranhos, SOD1, more than just an antioxidant, Archives of biochemistry and biophysics 697 (2021) 108701.
[25]. J. Niwa, S. Ishigaki, N. Hishikawa, M. Yamamoto, M. Doyu, S. Murata, K. Tanaka, N. Taniguchi, G. Sobue, Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity, The Journal of biological chemistry 277(39) (2002) 36793-8.
[26]. L.J. Martin, S.J. Koh, A. Price, D. Park, B.W. Kim, Nuclear Localization of Human SOD1 in Motor Neurons in Mouse Model and Patient Amyotrophic Lateral Sclerosis: Possible Links to Cholinergic Phenotype, NADPH Oxidase, Oxidative Stress, and DNA Damage, International journal of molecular sciences 25(16) (2024).
Cite this article
Emhenya,S.O. (2025). Screening Possible Substrates of E3 Ligase RNF19A by Immunofluorescence Staining. Theoretical and Natural Science,139,17-27.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. M.S. Moran, Radiation therapy in the locoregional treatment of triple-negative breast cancer, The Lancet. Oncology 16(3) (2015) e113-22.
[2]. A.N. Giaquinto, H. Sung, L.A. Newman, R.A. Freedman, R.A. Smith, J. Star, A. Jemal, R.L. Siegel, Breast cancer statistics 2024, CA: a cancer journal for clinicians 74(6) (2024) 477-495.
[3]. G. Bianchini, C. De Angelis, L. Licata, L. Gianni, Treatment landscape of triple-negative breast cancer - expanded options, evolving needs, Nature reviews. Clinical oncology 19(2) (2022) 91-113.
[4]. Y. Hu, C. Wang, H. Liang, J. Li, Q. Yang, The treatment landscape of triple-negative breast cancer, Medical oncology (Northwood, London, England) 41(10) (2024) 236.
[5]. A.M. Karim, J. Eun Kwon, T. Ali, J. Jang, I. Ullah, Y.G. Lee, D.W. Park, J. Park, J.W. Jeang, S.C. Kang, Triple-negative breast cancer: epidemiology, molecular mechanisms, and modern vaccine-based treatment strategies, Biochemical pharmacology 212 (2023) 115545.
[6]. S. Dawood, Triple-negative breast cancer: epidemiology and management options, Drugs 70(17) (2010) 2247-58.
[7]. I. Susumu, I. Kiyotaka, S. Shinichi, U. Keiji, H. Naoki, Y. Hiromasa, T. Yoshinori, Benefits of terminal noncardioplegic warm blood retrograde perfusion after terminal warm blood cardioplegia perfusion prior to aortic unclamping in open heart surgery, The Journal of cardiovascular surgery 47(6) (2006) 677-82.
[8]. D.H. Schlesinger, G. Goldstein, H.D. Niall, The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells, Biochemistry 14(10) (1975) 2214-8.
[9]. D.H. Schlesinger, G. Goldstein, Molecular conservation of 74 amino acid sequence of ubiquitin between cattle and man, Nature 255(5507) (1975) 423-4.
[10]. D. Popovic, D. Vucic, I. Dikic, Ubiquitination in disease pathogenesis and treatment, Nature medicine 20(11) (2014) 1242-53.
[11]. R. Kandel, J. Jung, S. Neal, Proteotoxic stress and the ubiquitin proteasome system, Seminars in cell & developmental biology 156 (2024) 107-120.
[12]. E. Rivkin, A.L. Kierszenbaum, M. Gil, L.L. Tres, Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head-tail coupling apparatus during rat spermatid development, Developmental dynamics : an official publication of the American Association of Anatomists 238(7) (2009) 1851-61.
[13]. C. Wu, Z. Su, M. Lin, J. Ou, W. Zhao, J. Cui, R.F. Wang, NLRP11 attenuates Toll-like receptor signalling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A, Nature communications 8(1) (2017) 1977.
[14]. Q. Zhu, J. Huang, H. Huang, H. Li, P. Yi, J.A. Kloeber, J. Yuan, Y. Chen, M. Deng, K. Luo, M. Gao, G. Guo, X. Tu, P. Yin, Y. Zhang, J. Su, J. Chen, Z. Lou, RNF19A-mediated ubiquitination of BARD1 prevents BRCA1/BARD1-dependent homologous recombination, Nature communications 12(1) (2021) 6653.
[15]. V.U. Bai, O. Hwang, G.W. Divine, E.R. Barrack, M. Menon, G.P. Reddy, C. Hwang, Averaged differential expression for the discovery of biomarkers in the blood of patients with prostate cancer, PloS one 7(4) (2012) e34875.
[16]. K. Im, S. Mareninov, M.F.P. Diaz, W.H. Yong, An Introduction to Performing Immunofluorescence Staining, Methods in molecular biology (Clifton, N.J.) 1897 (2019) 299-311.
[17]. H. Deng, G. Ji, J. Ma, J. Cai, S. Cheng, F. Cheng, RNF19A inhibits bladder cancer progression by regulating ILK ubiquitination and inactivating the AKT/mTOR signalling pathway, Biology direct 19(1) (2024) 102.
[18]. S. Zhu, M. Tao, Y. Li, X. Wang, Z. Zhao, Y. Liu, Q. Li, Q. Li, Y. Lu, Y. Si, S. Cao, J. Ye, H3K27me3 of Rnf19a promotes neuroinflammatory response during Japanese encephalitis virus infection, Journal of neuroinflammation 20(1) (2023) 168.
[19]. H. Zuo, J. Park, A. Frangaj, J. Ye, G. Lu, J.J. Manning, W.B. Asher, Z. Lu, G.B. Hu, L. Wang, J. Mendez, E. Eng, Z. Zhang, X. Lin, R. Grassucci, W.A. Hendrickson, O.B. Clarke, J.A. Javitch, A.D. Conigrave, Q.R. Fan, Promiscuous G-protein activation by the calcium-sensing receptor, Nature 629(8011) (2024) 481-488.
[20]. Q. Zhou, M.M. Zhang, M. Liu, Z.G. Tan, Q.L. Qin, Y.G. Jiang, LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson's disease progression, Aging 13(3) (2021) 4115-4137.
[21]. R. Mohammad Malyar, H. Li, H. Enayatullah, L. Hou, R. Ahmad Farid, D. Liu, J. Akhter Bhat, J. Miao, F. Gan, K. Huang, X. Chen, Zinc-enriched probiotics enhanced growth performance, antioxidant status, immune function, gene expression, and morphological characteristics of Wistar rats raised under high ambient temperature, 3 Biotech 9(8) (2019) 291.
[22]. Y. Cheng, Y. Hu, H. Wang, Z. Zhao, X. Jiang, Y. Zhang, J. Zhang, Y. Tong, X. Qiu, Ring finger protein 19A is overexpressed in non-small cell lung cancer and mediates p53 ubiquitin-degradation to promote cancer growth, Journal of cellular and molecular medicine 25(16) (2021) 7796-7808.
[23]. P. Cohen, M. Tcherpakov, Will the ubiquitin system furnish as many drug targets as protein kinases?, Cell 143(5) (2010) 686-93.
[24]. E.C.A. Eleutherio, R.S. Silva Magalhães, A. de Araújo Brasil, J.R. Monteiro Neto, L. de Holanda Paranhos, SOD1, more than just an antioxidant, Archives of biochemistry and biophysics 697 (2021) 108701.
[25]. J. Niwa, S. Ishigaki, N. Hishikawa, M. Yamamoto, M. Doyu, S. Murata, K. Tanaka, N. Taniguchi, G. Sobue, Dorfin ubiquitylates mutant SOD1 and prevents mutant SOD1-mediated neurotoxicity, The Journal of biological chemistry 277(39) (2002) 36793-8.
[26]. L.J. Martin, S.J. Koh, A. Price, D. Park, B.W. Kim, Nuclear Localization of Human SOD1 in Motor Neurons in Mouse Model and Patient Amyotrophic Lateral Sclerosis: Possible Links to Cholinergic Phenotype, NADPH Oxidase, Oxidative Stress, and DNA Damage, International journal of molecular sciences 25(16) (2024).









