

About JFSNHJournal of Food Science, Nutrition and Health (JFSNH) is an open-access, peer-reviewed academic journal hosted by Hubei Jingchu Specialty Food Industry Technology Research Institute and published by EWA Publishing. JFSNH is published irregularly. JFSNH present latest theoretical and methodological discussions to bear on the scholarly works covering food science, nutrition and health. Situated at the forefront of the interdisciplinary fields of food science and nutrition, this journal seeks to bring together the scholarly insights centring on food science & engineering, nutrition and health, chemical engineering, bioengineering and relevant subfields that trace to the discipline of food science, nutrition and chemical engineering, and combined fields of the aforementioned. JFSNH is dedicated to the gathering of intellectual views by scholars and policymakers. The articles included are relevant for scholars, policymakers, and students of food science & engineering, chemical engineering, bioengineering and otherwise interdisciplinary programs.For more details of the JFSNH scope, please refer to the Aim & Scope page. For more information about the journal, please refer to the FAQ page or contact info@ewapublishing.org. |
| Aims & scope of JFSNH are: ·Food Science & Engineering ·Nutrition ·Chemical Engineering ·Bioengineering |
Article processing charge
A one-time Article Processing Charge (APC) of 450 USD (US Dollars) applies to papers accepted after peer review. excluding taxes.
Open access policy
This is an open access journal which means that all content is freely available without charge to the user or his/her institution. (CC BY 4.0 license).
Your rights
These licenses afford authors copyright while enabling the public to reuse and adapt the content.
Peer-review process
Our blind and multi-reviewer process ensures that all articles are rigorously evaluated based on their intellectual merit and contribution to the field.
Editors View full editorial board

Wuhan, China
chenshuai@whu.edu.cn

Kuala Lumpur

Kuala Lumpur

Wuhan, China
houwenfu@whpu.edu.cn
Latest articles View all articles
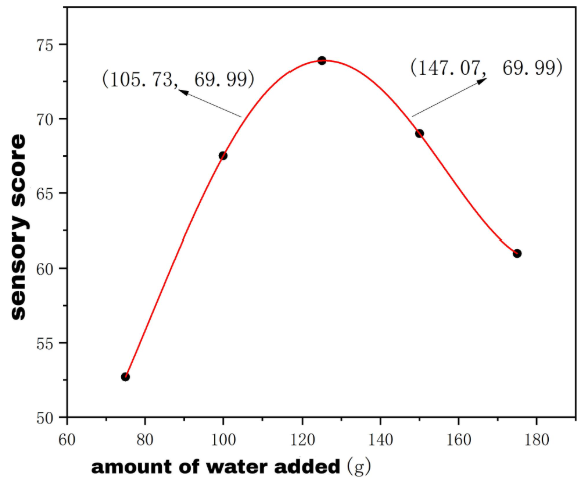
his study aimed to optimize the production process of Bazhen glutinous rice cake (Bazhen Nuomici) based on the concept of "medicinal food homology". The utilization efficiency of active ingredients in Bazhen herbal slices was improved by freeze-drying pretreatment combined with β-cyclodextrin inclusion technology. The response surface methodology was employed to optimize the raw material ratio, and a three-dimensional evaluation system integrating "process parameters-sensory quality-functional components" was established. Results indicated that the optimized product exhibited a 2.3-fold increase in astragaloside IV content (p<0.01), an 18.6% improvement in sensory score, and enhanced texture characteristics (hardness 120±5 g/cm², elasticity coefficient 0.85). The study provides technical insights into the functional enhancement of traditional glutinous rice products and the development of medicinal food homologues.

 View pdf
View pdf



Autism spectrum disorder (ASD) is increasingly prevalent, and evidence implicates gut–brain interactions, motivating evaluation of gut microbiome signals in ASD phenotypes. Prior studies link ASD with gastrointestinal comorbidity and dysbiosis, but taxonomic findings are heterogeneous across cohorts and methods, leaving uncertainty about which microbial features are most reproducible and informative. We analyzed two public pediatric cohorts (shotgun metagenomic and 16S rRNA) to assess cross-dataset separability of ASD versus neurotypical controls and to identify convergent microbial signals. After standard quality control and feature scaling, we trained logistic regression, random forest, and gradient boosting models, evaluated performance with confusion-matrix metrics and ROC-AUC, and compared taxa- and pathway-level importance to prioritize robust candidates. Models reliably distinguished ASD from controls, highlighting Prevotella and Bacteroides as consistent signals, with pathway analysis implicating short-chain fatty acid and tryptophan metabolism. Results support a biological role for microbiome dysbiosis in ASD, nominate reproducible genera and pathways as candidate biomarkers, and motivate longitudinal, strain-resolved, and interventional studies, including targeted probiotics or microbiota transfer, to test causality and therapeutic potential.

 View pdf
View pdf


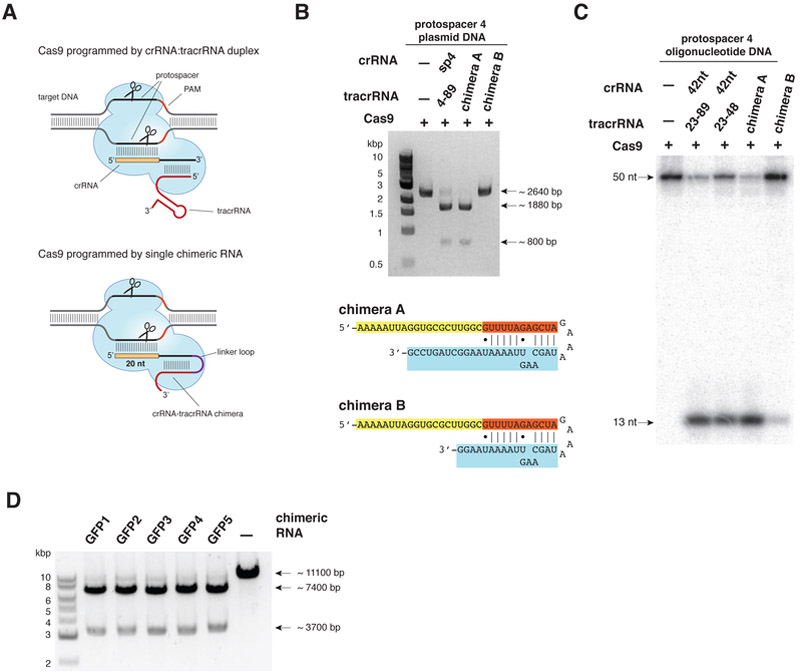
This paper reviews the latest advances and applications of gene editing technologies in the study of aging-related genes. In recent years, gene editing has achieved significant progress in the biomedical field, with continual improvements in accuracy and efficiency. Gene editing technologies demonstrate unique advantages in aging research, providing powerful tools for elucidating aging mechanisms and developing anti-aging interventions. This paper offers a detailed overview of major gene editing technologies (such as the CRISPR/Cas system, TALENs, and ZFNs) as well as emerging editing techniques (including base editing, prime editing, and epigenetic editing), describing their principles and applications. It also discusses research progress in areas such as constructing aging gene models, disease models, in vivo editing, and in vitro editing. Furthermore, it analyzes current technological challenges and proposes corresponding optimization strategies. Finally, it considers future directions for the development of gene editing technologies in aging research, including technological innovation and integrated multi-technology applications. Through these advances and innovations, gene editing is expected to play an increasingly important role in the anti-aging field, offering new strategies and methods to promote human health and longevity.

 View pdf
View pdf


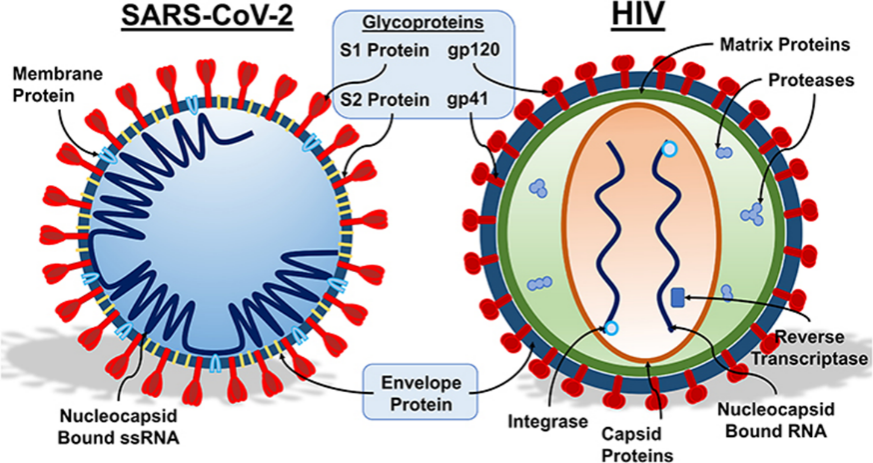
Viral antigen glycosylation is a central strategy for immune evasion. This review summarizes how viruses utilize glycosylation to form a "glycan shield" that occludes critical epitopes and to achieve molecular mimicry by imitating host "self" signals, thereby avoiding immune recognition. The article focuses on analyzing the challenges glycosylation poses to vaccine design, including heterogeneity, epitope masking, and dynamic evolution, as well as the obstacles it creates for antibody drugs, such as steric hindrance and drug resistance. Finally, it outlines future directions for overcoming this "glycan fortress" through cutting-edge analytical technologies, innovative intervention strategies, and artificial intelligence, offering insights for developing effective prevention and treatment strategies.

 View pdf
View pdf


Volumes View all volumes
2025
Volume 4September 2025
Find articlesVolume 4November 2025
Find articlesVolume 4November 2025
Find articles2024
Volume 2September 2024
Find articlesAnnouncements View all announcements
Journal of Food Science, Nutrition and Health
We pledge to our journal community:
We're committed: we put diversity and inclusion at the heart of our activities...
Journal of Food Science, Nutrition and Health
The statements, opinions and data contained in the journal Journal of Food Science, Nutrition and Health (JFSNH) are solely those of the individual authors and contributors...
Indexing
The published articles will be submitted to following databases below:





