1. Introduction
Staphylococcus aureus is a Gram-positive bacterium within the Bacillales order [1]. As one of the most common pathogens for humans, S. aureus expresses an extensive range of virulence factors, including toxins (haemolysins and leucocidins), immune evasive surface factors (e.g., capsule and protein A), and tissue invasion-promoting factors (e.g., hyaluronidase). Among these, many are acquired by mobile genetic elements, such as the most studied arginine-catabolic mobile element (ACME) and Paton-Valentine leucocidin (PVL) [2]. These virulence factors contribute to the wide range of S. aureus clinical infections, making it the most common cause of infective endocarditis [3] and three major types of osteoarticular infections [4], as well as important causes of bacteremia [5,6], skin and soft tissue infections, pneumonia [7] as well as other pleuropulmonary infections, and device-related infections [4].
The extensive and dynamic host range of S. aureus is also a widely researched aspect since its discovery, as the bacterium’s colonisation is observed in humans, livestock (e.g., cattle and lamb), companion animals (e.g., cats and dogs), and wild species [8,9].
However, perhaps the topic that gained S. aureus the most notice is a particular strain of the bacteria, namely, the methicillin-resistant staphylococcus aureus (MRSA). Bacteria gain antimicrobial resistance through simple modes of natural selection. Different levels of resistance to certain antibiotics constantly and spontaneously occur as the bacteria undergo mutations or horizontal gene transfers with other strains. After being administered to patients, the antibiotic places selective pressure on the bacterial strain. With sufficient time and prolonged antimicrobial treatment, selection eventually favours the strain that has acquired or accumulated the highest antibiotic minimum inhibitory concentrations (MICs) [2].
In the case of MRSA, S. aureus first demonstrated penicillin resistance in the 1940s [10] through the β-lactamase encoded by the blaZ gene. This gene was previously shown as a gene acquired by the transposon Tn552 or Tn552-like elements [11] and located on either a large plasmid or inserted into the bacterial chromatin [12]. After methicillin was introduced into clinical usage in 1959, methicillin-resistant S. aureus strains were discovered within two years in 1961 [13], the resistance rendered by the mecA gene acquired through the staphylococcus chromosome cassette mec (SCCmec) [14]. The SCCmec cassette contains the mecA gene complex and site-specific recombinase genes (ccrA and ccrB) responsible for methicillin resistance and mobility. It is integrated within the orfX gene in the S. aureus genome [15]. The mecA gene encodes for penicillin-binding protein 2a (PBP2a), a bacterial cell wall crossing peptidoglycan that demonstrates resistance to all β-lactam antibiotics due to its low affinity for β-lactams [16]. Since its first occurrence, MRSA has presented itself as a non-negligible problem for the medical system.
Apart from resistance to penicillin, methicillin, and other β-lactam antibiotics, MRSA also exhibits resistance to trimethoprim (acquired by gene dfrA and dfrK), erythromycin (developed by gene ermC), clindamycin (constitutively expressed ermC), tetracyclines (acquired by gene tetK and tetL) [2], and most recently, resistance to vancomycin, which was widely used as a treatment for MRSA before vancomycin-resistant S. aureus was observed in 1997 [17]. Thus, due to increased resistance to multiple antimicrobial drugs, MRSA is attributed to over 100,000 deaths and 3.5 million disability-adjusted life-years worldwide in 2019 [18], and 3.8% of the global population still carries the deadly pathogen [19].
Since the mecA gene is acquired and transmitted through a mobile genetic element that inserts itself into the host genome, the cassette must have originated from a separate organism other than S. aureus. The presence of the mecA gene and functional PBP2a protein has already determined in other bacterial species such as S. pseudintermedius and S. epidermidis (20,21). Apart from staphylococcal species, almost identical homologues of the mecA gene and PBP2a protein are also determined to be present in Mammaliicoccus sciuri (ex. Staphylococcus sciuri) and Mammaliicoccus fleurettii with high similarities, even able to fully replace the function of the mecA gene when inserted into an S. aureus bacterium [22,23]. The prevalence of the mecA gene and its corresponding PBP2a protein in multiple bacterial species could indicate an expansive range of methicillin and other β-lactam antibiotic resistance within the staphylococcal and mammalicoccal genus [24]. This research will focus on investigating the presence of the mecA gene and PBP2a protein among determined bacterial sequences, with a critical focus on analysing the structure and function of the PBP2a protein in multiple bacterial species.
2. Methods
To retrieve the mecA sequences, a search was carried out using the keywords “mecA” and “S. aureus” and the reference sequence of mecA (accession number MW682923). The corresponding amino acid sequence (accession number WP_000721310.1) Several data analyses using the Basic Local Alignment Search Tool (BLAST) were performed using different algorithms and exclusions. BLAST trails using BLASTn, tBLASTn, BLASTp, and BLASTx were performed using a query of either the mecA sequence or the PBP2a sequence. Within each database, four different trails adding different exclusions were simultaneously carried out, with the exclusion of none, Staphylococcus aureus (taxid 1280), Staphylococcus (taxid 1279), and Bacillus/Staphylococcus group (taxid 1385), respectively. To prevent counting in results generated by chance, bearing little biological significance, and to have poor similarity with the query sequence, at the end of each BLAST search, a filter is applied with percent identity 90%-100%, E-value 0-1e-6, and percent identity of 85%-100% was applied.
From BLASTn and BLASTx results, one PBP2a amino acid sequence from every Staphylococcal or Mammaliicoccal species is retrieved. Topological prediction using Phobius, annotation analysis and PDB structural file retrieving using Uniprot [25], conserved domain analysis using INTERPRO, and pairwise structural comparison using RCSB were carried out respectively using the amino acid sequences or the PDB structural files they correspond to.
3. Results
3.1. The mecA gene and PBP2a protein exist in multiple staphylococcus and mammaliicoccus species.
In the first BLASTn and tBLASTn search with no specific exclusions, aligned sequences came mainly from the S. aureus species. Apart from S. aureus, S. epidermidis also occupied a sizable portion of results under S. aureus, though S. epidermidis results coming from tBLASTn were significantly less than that coming from BLASTn. Both searches, excluding S. aureus (taxid 1280), yielded similar results, with aligned sequences mainly coming from S. epidermidis. Still, tBLASTn results included alignments from 4 species that were not included in BLASTn results. In the BLASTn and tBLASTn search with the exclusion of Staphylococcus (taxid 1279), both BLASTn and tBLASTn had the most results from M. sciuri. In BLASTn, there also existed results from four enterococcus species. In the search with the exclusion of Bacillus/Staphylococcus, valid results from BLASTn cover only the four enterococcus species, while tBLASTn yielded no significant results.
Table 1. BLAST analysis results

BLASTp and BLASTx search yielded identical results with any exclusion applied. In the investigation with no exclusion, most results came from S. aureus. From the search with the exclusion of S. aureus (taxid 1280), alignments came mainly from the species S. epidermidis and S. pseudintermedius. In the investigation with the exclusion of the staphylococcus group, results primarily originated from the species M. sciuri. Still, in both BLASTp and BLASTx, the search with the exclusion of the Bacillus/Staphylococcus group (taxid 1385) yielded no significant results (see table 1). The amino acid sequences of PBP2a protein from nine species with the most resemblance to the query were retrieved for further analysis (see table 2).
Table 2. Retrieved amino acid sequences of PBP2a protein from nine different bacterial species
Species | Accession number |
S.aureus | WP_000721310.1 |
S.epidermidis | MCC3680226.1 |
S.pseudintermedius | WP_140240747.1 |
S.argenteus | WP_088811536.1 |
S.haemolyticus | WP_080400705.1 |
S.hominis | OAW33239.1 |
S,cohnii | ADM43473.1 |
M.sciuri | WP_204254155.1 |
M.fleurettii | WP_203154062.1 |
3.2. Protein topology prediction suggests high conserveness of PBP2a in nine different staphylococcal and mammaliicoccal species.
FASTA sequences of the PBP2a protein from nine different staphylococcus/mammaliicoccus species (S. aureus, S. epidermidis, S. pseudintermedius, S. argenteus, S. haemolyticus, S. hominis, S. cohnii, M. sciuri, and M. fleurettii) were put into trials of transmembrane topology and signal peptide prediction tests. Results (as shown in fig 1) suggest that despite its presence in multiple different Staphylococcus and Mammaliicoccus, PBP2a protein shared similar protein signatures, all nine proteins bearing a transmembrane topology order of a cytoplasmic region with the length of 6aa, a transmembrane region with the length of 18-19aa, and a non-cytoplasmic region with a length of 644aa. The only minor difference in the protein signature between species is that the PBP2a protein sequences of S. epidermidis, S. pseudintermedius, S. argenteus, S. haemolyticus, M. sciuri, and M. fleurettii bear a transmembrane region with the length of 19aa, differing from that of the others whose same region is of 18aa in length.
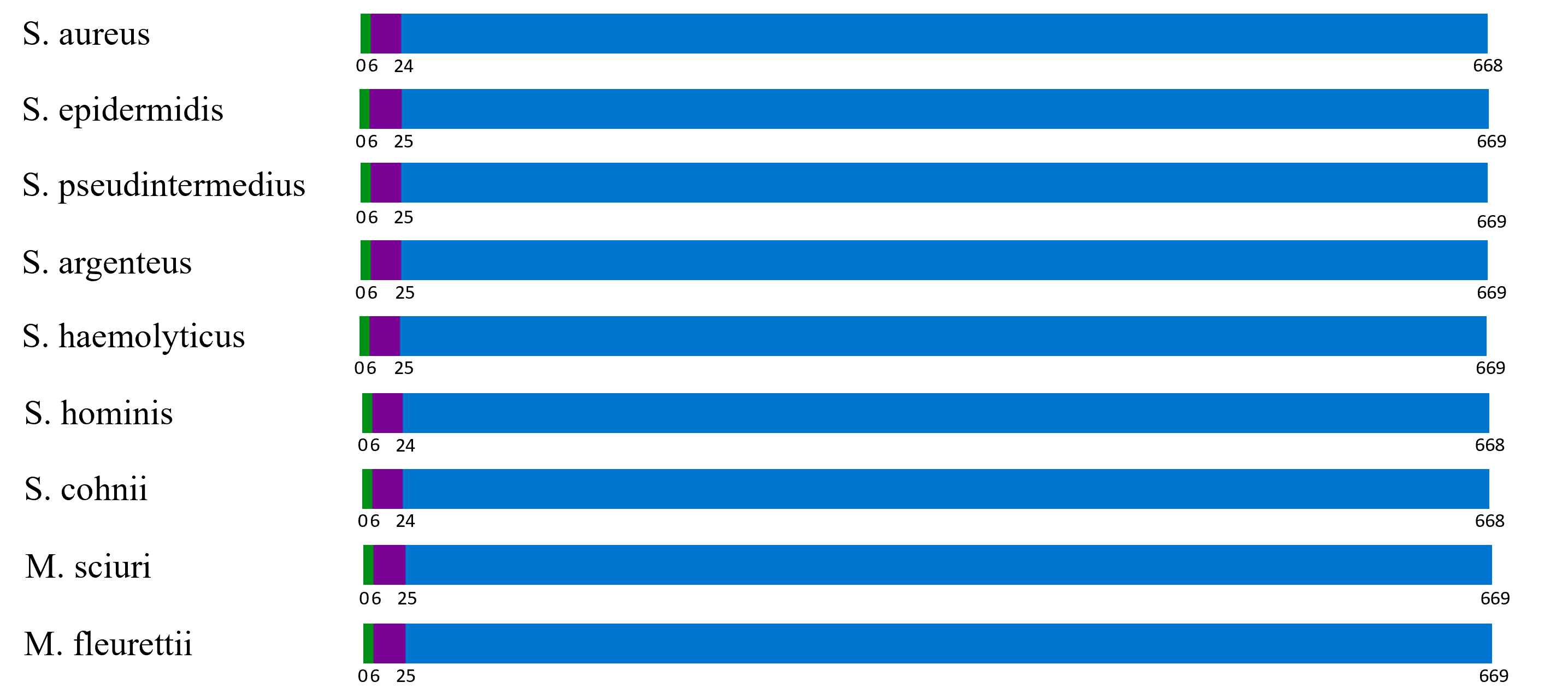
Figure 1. Phobius topology prediction of the cytoplasmic region (green), transmembrane region (purple), and non-cytoplasmic region (blue).
3.3. High similarities of conserved domains in PBP2a indicate similar functions between bacterial species
The FASTA sequences of the PBP2a protein originated from nine different bacterial species and were put into INTERPRO trails to determine conserved domains within the nine sequences. Results (as shown in fig 2) indicated that all nine sequences share similar conserved domains. In the non-cytoplasmic regions of the nine proteins, the conserved domains mecA_N (115aa in length), PBP dimer (162aa in size), and PCN binding transpeptidase (312aa in length).
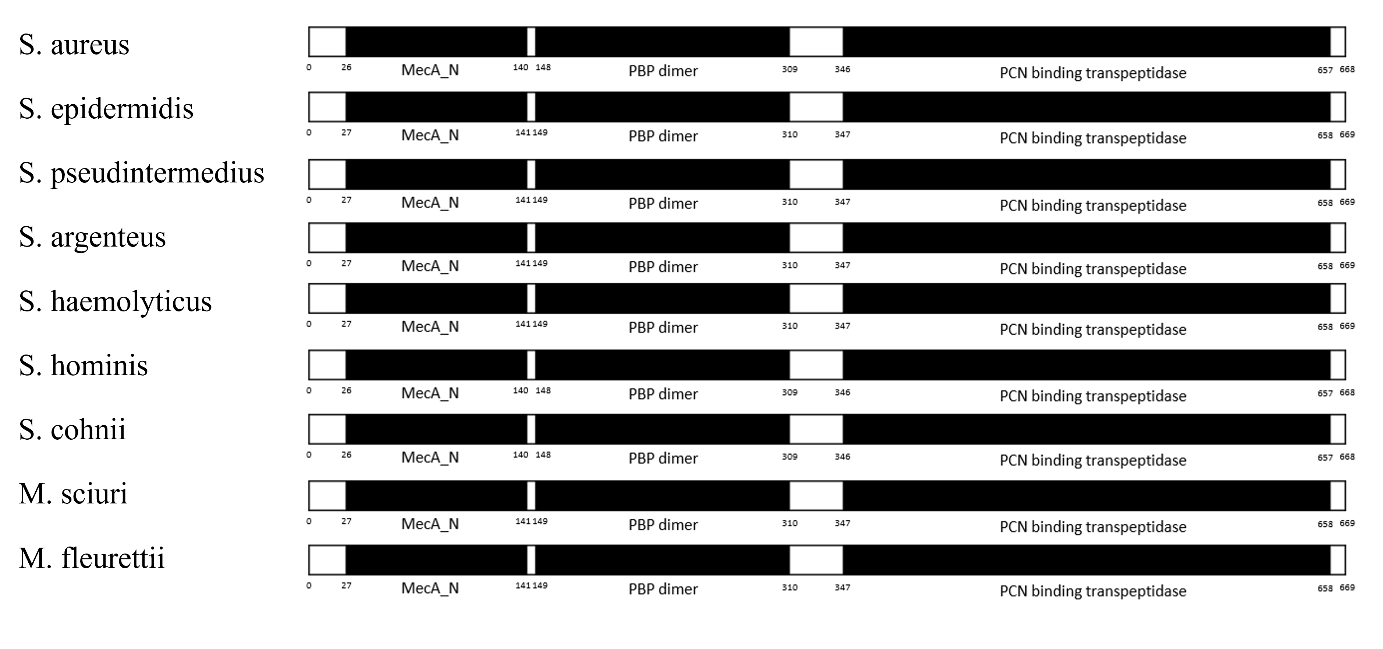
Figure 2. A comparison of the INTERPRO conserved domain analysis of PBP2a from nine different species.
4. Discussion
From BLAST search analysis, one may conclude that the mecA gene and PBP2a protein is somewhat prevalent in staphylococcal and mammaliicoccal genus. Apart from S. aureus, the mecA gene was also observed in 12 different staphylococcal bacterial species and four different mammaliicoccal species. These results indicate that many species in the two groups have been observed to bear the mecA gene in their genome. An explanation for such high prevalence. However, despite the high prevalence of the mecA gene, the PBP2a protein it codes for is somewhat less common in those species; only in a mere seven staphylococcal species and two mammaliicoccal species were homologous proteins observed in this study. This might be explained by the lack of data for PBP2a protein except for the few species (such as S. aureus, S. epidermidis, M. sciuri, etc.) where the most significant research takes place.
Apart from traces of the existence of the mecA gene and PBP2a protein in staphylococcal and mammaliicoccal bacterial species, BLASTn also suggested a nucleotide sequence similar to that of mecA in S. aureus exists in three enterococcus species, one mycobacterium species, and one proteus species. Even so, homologous proteins to PBP2a were not found in corresponding BLASTp and BLASTx searches. This might be explained as horizontal gene transfer that occurred purely by chance, with six foreign genuses acquiring the mecA gene through mobile genetic elements but unable to express them.
From Phobius topology prediction, conserved domain analysis, and protein 3-D structure comparison, it is of great significance that the PBP2a protein is highly conserved among different staphylococcal and mammaliicoccal bacterial species. From the analysis of 7 staphylococcal species and two mammaliicoccal species, it is clear that structure and domain function is identical among these species. This phenomenon is of conservation among antimicrobial genes and proteins and is not without logic. From the Phobius topological and conserved domain analysis, it can be concluded that a majority of the PBP2a protein is outside of the cytomembrane, and all the conserved domains are located within the non-cytoplasmic region of the protein, presumably to fulfil its function of deactivating the antimicrobial agent before it enters the cell. As long as the ligand, in this case, methicillin and other β lactam antibiotics do not change its structure, so would not the structure of the protein.
In the results collecting section from BLAST, it is easily noticed that despite BLASTp and BLASTx yielding identical results, BLASTn and tBLASTn generated more dissimilar results. This could be partly attributed to the differences in the translation algorithm between nucleotide to amino acids and adversely, for it is commonly known that one specific amino acid most likely corresponds to multiple codons. At the same time, the several-for-one pattern does not necessarily apply inversely.
5. Conclusion
BLAST results from this study indicated that both the mecA gene and its corresponding PBP2a protein are present and even of noticeable prevalence beyond S. aureus, with the presence of mecA spreading into other species of Staphylococci: S. epidermidis, S. pseudintermedius, S. argenteus, S. haemolyticus, S. hominis, S. hyicus, S. capitis, S. warneri, S. lugdunensis, S. taiwanensis, S. schleiferi, S. caprae, and into the Mammaliicoccus genus: M. sciuri, M. vitulinus, M. lentus, and M. fleurettii. Similarly, the PBP2a protein is also present in both groups, including BLAST hits in S. epidermidis, S. pseudintermedius, S. argenteus, S. haemolyticus, S. hominis, S. cohnii, M. sciuri, M. vitulinus, M. fleurettii, and M. massiliense. Furthermore, of the species bearing the PBP2a protein, the topology, domains, functions, and structure are highly conserved and highly similar to that of S. aureus.
References
[1]. Gnanamani A, Hariharan P, Paul-Satyaseela M. Staphylococcus aureus: Overview of Bacteriology, Clinical Diseases, Epidemiology, Antibiotic Resistance and Therapeutic Approach. 2017. Frontiers in S. aureus, InTech; https://doi.org/10.5772/67338
[2]. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. 2019 Nat. Rev. Microbiol. 17 203–18. https://doi.org/10.1038/s41579-018-0147-4
[3]. Fowler VG, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, et al. Staphylococcus aureus Endocarditis A Consequence of Medical Progress. 2005 JAMA 293(24) 3012–3021. https://doi.org/10.1001/jama.293.24.3012
[4]. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. 2015 Clin. Microbiol Rev. 28 603–61. https://doi.org/10.1128/CMR.00134-14
[5]. Hassoun A, Linden PK, Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. 2017 Crit. Care 21 211. https://doi.org/10.1186/s13054-017-1801-3
[6]. Pastagia M, Kleinman LC, de la Cruz EGL, Jenkins SG. Predicting risk for death from MRSA bacteremia. 2012 Emerg. Infect. Dis. 18 1072–80. https://doi.org/10.3201/eid1807.101371
[7]. American Thoracic Society. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. 2005 Am. J. Respir. Crit. Care Med 171 388–416. https://doi.org/10.1164/rccm.200405-644ST
[8]. Matuszewska M, Murray GGR, Harrison EM, Holmes MA, Weinert LA. The Evolutionary Genomics of Host Specificity in Staphylococcus aureus. 2020 Trends Microbiol. 28 465–77. https://doi.org/10.1016/j.tim.2019.12.007
[9]. Lee J H. Occurrence of methicillin-resistant Staphylococcus aureus strains from cattle and chicken, and analyses of their mecA, mecR1 and mecI genes. 2006 Vet. Microbiol. 114 155–9. https://doi.org/10.1016/j.vetmic.2005.10.024
[10]. Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. 2003 J. of Clinical Investigation 111 1265–73. https://doi.org/10.1172/jci200318535
[11]. Jensen SO, Lyon BR. Genetics of antimicrobial resistance in Staphylococcus aureus. 2009 Future Microbiol. 4 565–82. https://doi.org/10.2217/fmb.09.30
[12]. Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. 2017 FEMS Microbiol. Rev. 41 430–49. https://doi.org/10.1093/femsre/fux007
[13]. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). 2002 Proc. Natl. Acad. Sci. USA 99.; 7687–7692
[14]. Katayama Y, Ito T, Hiramatsu K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. 2000 Antimicrobial agents and chemotherapy 44 1549–1555
[15]. Ito T, Kuwahara K, Hiramatsu K. Staphylococcal Cassette Chromosome mec (SCCmec) Analysis of MRSA. 2013 Methods of Mol. Bio. 1085 131–148
[16]. Hartman BJ, Tomasz A, Sabath L. Low-Affinity Penicillin-Binding Protein Associated with r-Lactam Resistance in Staphylococcus aureus methicillin-resistant strain Col was supplied. 1984 J. Bacteriol. 158.
[17]. Gardete S, Tomasz A. Mechanisms of vancomycin resistance in Staphylococcus aureus. 2014 J. Clinical Investigation 124 2836–40. https://doi.org/10.1172/JCI68834.
[18]. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. 2022 The Lancet 399 629–55. https://doi.org/10.1016/S0140-6736(21)02724-0
[19]. Williamson K, Bisaga A, Paquette K, Lovell E. The prevalence of methicillin-resistant Staphylococcus aureus colonization in emergency department fast track patients. 2013 World J. Emerg. Med. 4 278. https://doi.org/10.5847/wjem.j.issn.1920-8642.2013.04.006
[20]. Najar-Peerayeh S, Moghadas AJ, Behmanesh M. Antibiotic susceptibility and mecA frequency in staphylococcus epidermidis, isolated from intensive care unit patients. 2014 Jundishapur J. Microbiol. 7(8) https://doi.org/10.5812/jjm.11188
[21]. Gagetti P, Errecalde L, Wattam AR, De Belder D, Ojeda Saavedra M, Corso A, et al. Characterization of the First mecA-Positive Multidrug-Resistant Staphylococcus pseudintermedius Isolated from an Argentinian Patient. 2020 Microbial Drug Resistance 26 717–21. https://doi.org/10.1089/mdr.2019.0308.
[22]. Fuda C, Suvorov M, Shi Q, Hesek D, Lee M, Mobashery S. Shared functional attributes between the mecA gene product of Staphylococcus sciuri and penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. 2007 Biochemistry 46 8050–7. https://doi.org/10.1021/bi7004587.
[23]. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. 2010 Antimicrob. Agents Chemotherapy 54 4352–9. https://doi.org/10.1128/AAC.00356-10.
[24]. Gill SR, Fouts DE, Archer GL, Mongodin EF, DeBoy RT, Ravel J, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. 2005 J. Bacteriol. 187 2426–38. https://doi.org/10.1128/JB.187.7.2426-2438.2005.
[25]. Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, et al. UniProt: the universal protein knowledgebase in 2021 2021 Nucleic Acids Res. 49 480–9. https://doi.org/10.1093/nar/gkaa1100.
Cite this article
Liu,Y. (2024). Investigating the prevalence and function of the mecA gene and PBP2a protein among Staphylococcus and Mammaliicoccus species. Theoretical and Natural Science,35,119-125.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Gnanamani A, Hariharan P, Paul-Satyaseela M. Staphylococcus aureus: Overview of Bacteriology, Clinical Diseases, Epidemiology, Antibiotic Resistance and Therapeutic Approach. 2017. Frontiers in S. aureus, InTech; https://doi.org/10.5772/67338
[2]. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. 2019 Nat. Rev. Microbiol. 17 203–18. https://doi.org/10.1038/s41579-018-0147-4
[3]. Fowler VG, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, et al. Staphylococcus aureus Endocarditis A Consequence of Medical Progress. 2005 JAMA 293(24) 3012–3021. https://doi.org/10.1001/jama.293.24.3012
[4]. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. 2015 Clin. Microbiol Rev. 28 603–61. https://doi.org/10.1128/CMR.00134-14
[5]. Hassoun A, Linden PK, Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. 2017 Crit. Care 21 211. https://doi.org/10.1186/s13054-017-1801-3
[6]. Pastagia M, Kleinman LC, de la Cruz EGL, Jenkins SG. Predicting risk for death from MRSA bacteremia. 2012 Emerg. Infect. Dis. 18 1072–80. https://doi.org/10.3201/eid1807.101371
[7]. American Thoracic Society. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. 2005 Am. J. Respir. Crit. Care Med 171 388–416. https://doi.org/10.1164/rccm.200405-644ST
[8]. Matuszewska M, Murray GGR, Harrison EM, Holmes MA, Weinert LA. The Evolutionary Genomics of Host Specificity in Staphylococcus aureus. 2020 Trends Microbiol. 28 465–77. https://doi.org/10.1016/j.tim.2019.12.007
[9]. Lee J H. Occurrence of methicillin-resistant Staphylococcus aureus strains from cattle and chicken, and analyses of their mecA, mecR1 and mecI genes. 2006 Vet. Microbiol. 114 155–9. https://doi.org/10.1016/j.vetmic.2005.10.024
[10]. Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. 2003 J. of Clinical Investigation 111 1265–73. https://doi.org/10.1172/jci200318535
[11]. Jensen SO, Lyon BR. Genetics of antimicrobial resistance in Staphylococcus aureus. 2009 Future Microbiol. 4 565–82. https://doi.org/10.2217/fmb.09.30
[12]. Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. 2017 FEMS Microbiol. Rev. 41 430–49. https://doi.org/10.1093/femsre/fux007
[13]. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). 2002 Proc. Natl. Acad. Sci. USA 99.; 7687–7692
[14]. Katayama Y, Ito T, Hiramatsu K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. 2000 Antimicrobial agents and chemotherapy 44 1549–1555
[15]. Ito T, Kuwahara K, Hiramatsu K. Staphylococcal Cassette Chromosome mec (SCCmec) Analysis of MRSA. 2013 Methods of Mol. Bio. 1085 131–148
[16]. Hartman BJ, Tomasz A, Sabath L. Low-Affinity Penicillin-Binding Protein Associated with r-Lactam Resistance in Staphylococcus aureus methicillin-resistant strain Col was supplied. 1984 J. Bacteriol. 158.
[17]. Gardete S, Tomasz A. Mechanisms of vancomycin resistance in Staphylococcus aureus. 2014 J. Clinical Investigation 124 2836–40. https://doi.org/10.1172/JCI68834.
[18]. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. 2022 The Lancet 399 629–55. https://doi.org/10.1016/S0140-6736(21)02724-0
[19]. Williamson K, Bisaga A, Paquette K, Lovell E. The prevalence of methicillin-resistant Staphylococcus aureus colonization in emergency department fast track patients. 2013 World J. Emerg. Med. 4 278. https://doi.org/10.5847/wjem.j.issn.1920-8642.2013.04.006
[20]. Najar-Peerayeh S, Moghadas AJ, Behmanesh M. Antibiotic susceptibility and mecA frequency in staphylococcus epidermidis, isolated from intensive care unit patients. 2014 Jundishapur J. Microbiol. 7(8) https://doi.org/10.5812/jjm.11188
[21]. Gagetti P, Errecalde L, Wattam AR, De Belder D, Ojeda Saavedra M, Corso A, et al. Characterization of the First mecA-Positive Multidrug-Resistant Staphylococcus pseudintermedius Isolated from an Argentinian Patient. 2020 Microbial Drug Resistance 26 717–21. https://doi.org/10.1089/mdr.2019.0308.
[22]. Fuda C, Suvorov M, Shi Q, Hesek D, Lee M, Mobashery S. Shared functional attributes between the mecA gene product of Staphylococcus sciuri and penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. 2007 Biochemistry 46 8050–7. https://doi.org/10.1021/bi7004587.
[23]. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. 2010 Antimicrob. Agents Chemotherapy 54 4352–9. https://doi.org/10.1128/AAC.00356-10.
[24]. Gill SR, Fouts DE, Archer GL, Mongodin EF, DeBoy RT, Ravel J, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. 2005 J. Bacteriol. 187 2426–38. https://doi.org/10.1128/JB.187.7.2426-2438.2005.
[25]. Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, et al. UniProt: the universal protein knowledgebase in 2021 2021 Nucleic Acids Res. 49 480–9. https://doi.org/10.1093/nar/gkaa1100.









