1. Introduction
COVID-19 represents a worldwide public health emergency triggered by the SARS-CoV-2 virus, which rapidly spread worldwide, leading to millions of infections and Governments and international organizations have implemented various measures to limit the transmission of the virus, including travel restrictions, home quarantines, social distancing, and mask-wearing. Although vaccination efforts have been somewhat successful in reducing severe cases and mortality, the emergence of virus variants continues to challenge containment efforts. COVID-19 has not only had profound negative effects on the global economy and society but has also exacerbated social inequalities and mental health issues while increasing pressure on public health systems. This crisis has highlighted the vulnerabilities of global health systems and the importance of international cooperation.
In the fight against COVID-19, global public health systems have faced numerous challenges, including the virus's high transmissibility, the scarcity of medical resources, and the uncertainty of information. In this context, efficient diagnostic tools are of paramount importance. While PCR, antigen, and antibody tests are the primary diagnostic methods, they are hindered by certain complexities and delays, limiting their efficiency and timeliness. In contrast, chest CT scans can provide more detailed lung images, clearly showing subtle changes in lung lesions, making them suitable for early detection and disease monitoring. CT imaging can not only quickly identify lung manifestations in infected individuals but also more accurately assess the progression of the disease. Therefore, finding fast and accurate diagnostic methods, particularly those that utilize CT imaging analysis, has become an urgent priority.
Traditional medical imaging analysis methods mainly rely on radiologists to manually evaluate CT images. These approaches are not only time-intensive but also vulnerable to subjective influences, which may result in inconsistencies and inaccuracies in diagnosis. However, with computer vision technology developing rapidly, automated image analysis has gradually shown great potential. As a crucial branch of artificial intelligence, computer vision processes images automatically, allowing valuable information to be quickly extracted, thereby assisting physicians in making more accurate diagnoses and decisions.
Convolutional neural networks (CNNs) have emerged as the central technology for image classification tasks within the domain of deep learning. Inspired by the human visual system, CNNs utilize several layers of convolutional operations to automatically derive features from images, enabling efficient image content classification. Early CNN models like LeNet-5 and AlexNet significantly improved image recognition accuracy by introducing multiple layers of convolution and pooling operations [1][2]. In recent years, ResNet has emerged as a crucial deep learning architecture by introducing residual connections that effectively address the vanishing gradient problem in deep network training, significantly improving classification performance. The innovation of ResNet lies in its skip connections, allowing the network to directly learn residuals, thereby achieving deeper feature identification and categorization.
The continuous advancement of deep learning has led to the remarkable performance of convolutional neural networks (CNNs) in the field of image processing. Particularly in COVID-19 image recognition, the pre-trained ResNet model has demonstrated its significant promise in automated analysis of medical images through efficient feature extraction and accurate classification. By applying deep learning technology to CT image classification, this study aims to utilize the pre-trained ResNet model to enhance the speed and precision of COVID-19 diagnosis, alleviate the workload of physicians, and provide valuable insights for image analysis in future public health crises.
2. Previous works
Medical image classification is a significant research area in medical informatics. Achieving automated medical image classification is a key component of intelligent medical diagnostic technologies. With the swift progress of medical imaging technology, a range of machine learning methods have been extensively employed for the automated classification and diagnosis of medical images, aiming to enhance diagnostic accuracy and efficiency. Traditional image classification methods mainly include logistic regression, support vector machines (SVM), decision trees, and naive Bayes classifiers.
Logistic regression, a simple and efficient statistical model, is primarily used for predicting binary classification outcomes. Its advantage lies in its computational efficiency. however, due to its linear nature, logistic regression struggles when handling complex patterns in medical images [3]. Support vector machines (SVM), based on supervised learning algorithms, achieve linear or nonlinear classification by selecting hyperplanes with the maximum margin. SVMs are known for their high accuracy and ability to handle small samples and high-dimensional data effectively, but they exhibit high computational complexity when dealing with large-scale data [4]. Decision trees are non-parametric machine learning models that classify and regress by partitioning the feature space. They are characterized by good interpretability, ease of visualization, and the ability to capture nonlinear relationships, yet they are prone to overfitting with smaller datasets [5]. The Naive Bayes classifier, which relies on Bayes' theorem, is computationally simple and efficient, maintaining low computational complexity even with high-dimensional inputs. However, its assumption of conditional independence among features may not hold in real-world applications, affecting classification performance [6].
Although traditional machine learning methods have played a significant role in medical image classification, they face numerous challenges. These methods typically rely on manual feature extraction, making it difficult to automatically learn complex spatial features and contextual information from images. Traditional methods assume that data are linearly separable, making it difficult to capture the nonlinear patterns and higher-level features present in medical images. Additionally, the performance of traditional machine learning models is limited by their model complexity and ability to handle high-dimensional data [7]. For example, logistic regression and naive Bayes models, due to their simple structure, are ineffective in processing the complex patterns in medical images. While SVMs and decision trees offer more flexibility, they still face limitations when handling large-scale, high-dimensional, and complexly structured data.
Compared to traditional methods, deep learning models, CNNs, offer notable advantages in the classification of medical images. CNNs are capable of autonomously learning and extracting spatial hierarchical features and complex patterns from images, thereby minimizing the reliance on manual feature extraction. These models abstract image features layer by layer through a multi-layer structure, allowing them to capture subtle differences and higher-level semantic information in medical images. This ability to automatically learn features and extract deep features has enabled deep learning models to excel in large-scale medical image classification tasks, significantly enhancing classification performance [8].
3. Dataset and Preprocessing
The dataset used in this study is the COVID-19 Radiography Database, provided by Tawsifur Rahman and his colleagues and made publicly available on the Kaggle platform. The dataset consists of a set of chest X-rays classified into four categories: COVID-19, Normal, Viral Pneumonia, and Lung Opacity, with specific sample counts of 3,616, 10,192, 1,345, and 6,012 images, respectively. The image resolutions vary, with the majority being 299×299 pixels and 256×256 pixels [9-10].
A systematic preprocessing was performed to make the data appropriate for training and evaluating machine learning models. Initially, all images were resized to a consistent resolution of 224×224 pixels. This resizing was aimed at standardizing the input dimensions, reducing computational overhead, and simplifying the model architecture. Next, to enhance model training stability and accelerate the convergence of the gradient descent process, the pixel values of the images were normalized to the range [0, 1], by dividing each pixel value by 255. Finally, the dataset was divided into training and validation sets in an 80:20 ratio, ensuring that the model was trained on a diverse dataset and its generalization ability could be effectively evaluated.
4. Model
In this study, we employed a CNN based on the ResNet50 architecture for categorizing COVID-19 radiographic images. The ResNet50 model, pre-trained on the ImageNet dataset, was used as the base network. This pre-training enabled the model to acquire comprehensive image features, enhancing its performance on specific tasks. To adapt to our dataset, the model's input layer was set to an image size of 128×128×3 to ensure consistency in the input data and meet the network's computational requirements. Specifically, we retained the convolutional layers of ResNet50 but removed the top classification layer to allow for further fine-tuning on the COVID-19 radiographic image dataset. To improve classification performance, we added a global average pooling layer, a fully connected layer, and an output layer on top of ResNet50. The global average pooling layer was employed to condense the spatial dimensions of the feature maps into a single value, which helped decrease the number of parameters and reduce computational complexity; The fully connected layer was configured with 1,024 neurons using a ReLU activation function, while the output layer consisted of 4 neurons and utilized the Softmax function for multi-class classification. The detailed model architecture is presented in Table 1.
Table 1. Model Framework.
Layer | Name | number of parameters |
1 | Resnet50 | 23,587,712 |
2 | GlobalAveragePooling2D | 0 |
3 | Dense (1024 units, ReLU) | 1,049,600 |
4 | Dropout (0.5) | 0 |
5 | Output Layer (4 units, Softmax) | 4,100 |
The model underwent training for 100 epochs, with early stopping implemented to avoid overfitting. In this study, categorical cross-entropy served as the loss function, which is especially appropriate for multi-class classification tasks, focusing on reducing the disparity between the predicted and actual class distributions. The loss function is calculated as:
\( Loss=-\sum _{i=1}^{N}{y_{i}}log({\hat{y}_{i}}) \)
where N is the number of classes, \( {y_{i}} \) is the indicator variable for the true class, and \( {\hat{y}_{i}} \) is the model's predicted probability that the sample belongs to class i. To accelerate the convergence process and adjust the learning rate, the Adam optimizer was employed with the learning rate adjusted to 0.00001.
5. Results
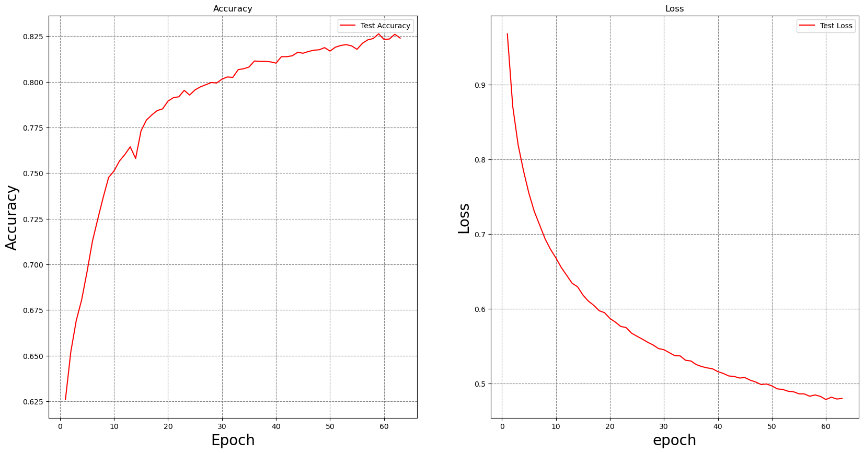
Figure 1. Accuracy and Loss Curves.
Figure 1 (left) shows the variation in test accuracy across training epochs. As depicted, the model's accuracy rapidly improves during the initial epochs, rising from approximately 62.5% to nearly 82.5%. This rapid growth in accuracy indicates that the model is successfully capturing the image features and gradually adapting to the dataset. As training continues, the rate of accuracy improvement slows down, and the accuracy stabilizes after around the 50th epoch. This suggests that the model has mostly converged, and further increasing the number of epochs does little to enhance accuracy. Ultimately, the model's accuracy stabilizes at around 82.33%, demonstrating good performance in the task of classifying COVID-19 radiographic images. Figure 1 (right) illustrates the trend in test loss over the course of training. It can be observed that the loss decreases rapidly in the early stages of training, dropping from nearly 1.0 to below 0.5. This indicates that the model is effectively optimizing its parameters and reducing prediction error. With the number of epochs increasing, the rate of loss reduction slows down, and the loss stabilizes after around the 50th epoch, indicating that the model has gradually converged, with no significant signs of overfitting.
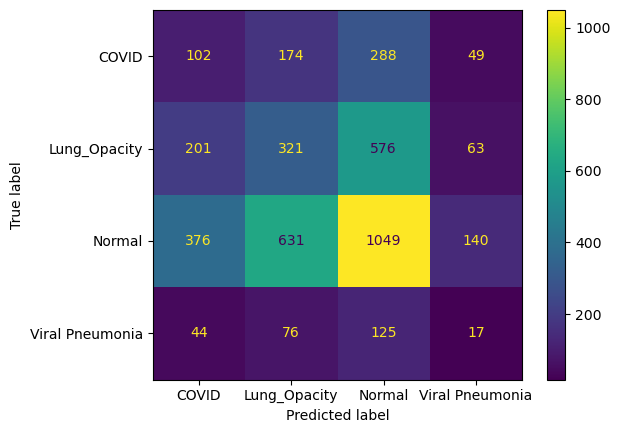
Figure 2. Confusion Matrix.
Figure 2 presents the confusion matrix for the validation set, which evaluates the model's classification performance across different categories. From the figure, it can be seen that the model's performance in the COVID category is less than satisfactory. Although it correctly classified 102 COVID images, a considerable number of COVID images were misclassified into other categories: 174 were misclassified as Lung Opacity, 288 as Normal, and 49 as Viral Pneumonia. This indicates that the model faces significant challenges in identifying COVID images, particularly in confusing them with Normal and Lung Opacity categories. For the Lung Opacity category, the model correctly classified 321 images, but a substantial number of samples were misclassified. Notably, 576 Lung Opacity images were misclassified as Normal, 201 as COVID, and 63 as Viral Pneumonia. This suggests that the model has some difficulty distinguishing between Lung Opacity and Normal images, likely due to similarities in their radiographic features. In the Normal category, the model performed relatively well, correctly classifying 1,049 samples. However, 631 Normal images were misclassified as Lung Opacity, 376 as COVID, and 140 as Viral Pneumonia. This indicates that, despite a higher correct classification rate for the Normal category, Normal images are still prone to misclassification, especially as Lung Opacity, which may reflect overlapping features between these two categories. The classification performance for the Viral Pneumonia category was relatively poor, with the model correctly classifying only 17 samples, while a large number of Viral Pneumonia images were misclassified into other categories: 44 as COVID, 76 as Lung Opacity, and 125 as Normal. This suggests that the model struggles to accurately identify Viral Pneumonia images, particularly tending to confuse them with Normal images.
6. Discussion and Conclusion
This study successfully constructed an efficient CNN for categorizing COVID-19 radiographic images by utilizing a pre-trained ResNet50 model and transfer learning technique. Experimental results indicated that the model attained an accuracy of 82.33% on the test set, validating its potential application in COVID-19 diagnosis. The model effectively distinguishes COVID-19 positive cases from other types of lung images, providing a reliable auxiliary diagnostic tool for medical professionals.
Despite the significant achievements of this study, several limitations need further exploration and resolution. First, the size and diversity of the dataset remain important constraints. The dataset used in this study is relatively limited in scale and primarily focuses on specific types of radiographic images. Subsequent research should focus on assembling larger and more varied datasets to more comprehensively validate the model's generalization ability and robustness, ensuring its applicability across different clinical scenarios. Second, although the ResNet50 model demonstrated excellent performance, its high computational complexity and significant demand for computing resources may limit its application in resource-constrained environments. Future studies might consider adopting more lightweight model architectures, such as MobileNet, to reduce computational costs and enable broader applications, particularly in mobile devices or telemedicine contexts. Additionally, while the current CNN model can provide highly accurate classification results, its internal workings are intricate and lack transparency, which makes it challenging for medical professionals to completely trust the model. Future research should integrate Explainable AI (XAI) methods to enhance the transparency and trustworthiness of the model, thereby increasing its acceptance in clinical practice. Addressing these limitations, future research should focus on expanding dataset diversity, model simplification, and improving interpretability to promote the widespread application and practical deployment of CNNs in the automatic categorization of COVID-19 radiographic images.
Based on this research, future work can be optimized and expanded in the following areas to further advance the use of deep learning models in COVID-19 and other medical image analyses. First, the diversity and scale of the dataset are key to further improving the model's generalization ability. Future research should collect more diverse large-scale datasets that cover different regions, populations, and healthcare institutions, which will not only help improve the model's robustness but also ensure its applicability and accuracy on a global scale. Meanwhile, with the emergence of new viral variants and diseases, continuous updates to the dataset will also be necessary. Second, the light weighting and efficiency of the model are important directions for future development. Although the ResNet50 model has demonstrated excellent performance, its high computational complexity limits its application in resource-constrained environments. Future efforts could explore the use of more lightweight models, such as MobileNet or EfficientNet, to reduce computational costs, enabling real-time applications of the model on mobile devices or in telemedicine systems. Techniques such as model quantization and pruning can further alleviate the computational burden. Regarding model interpretability, while current deep learning models are highly accurate, their "black-box" nature remains a challenge. Future research should integrate Explainable AI (XAI) technologies to make the decision-making process of the model more transparent through methods like visualization and attention mechanisms. This transparency will help physicians better understand the basis for the model's predictions, thereby increasing its acceptance in clinical practice. The integration of multimodal data is another promising area for future exploration. Current research mainly relies on single imaging data, but future studies could consider combining other types of medical data, such as electronic health records (EHR) and laboratory test results. This multimodal data fusion would provide more comprehensive information for disease diagnosis, thus improving the accuracy and reliability of the diagnoses. Finally, clinical validation and application are critical steps in translating research findings into practical benefits. Future research should involve multicenter clinical trials to validate the model's effectiveness in real-world settings and develop user-friendly clinical tools that allow physicians to easily incorporate deep learning models into routine diagnostic workflows. This would help increase diagnostic efficiency, improve patient care quality, and promote the widespread adoption of deep learning technologies in the healthcare sector. By exploring these research avenues and making corresponding enhancements, the application prospects of deep learning models in COVID-19 and other medical image analyses will be even more promising, contributing significantly to the advancement of healthcare.
References
[1]. Y. LeCun, L. Bottou, Y. Bengio, and P. Haffner, "Gradient-based learning applied to document recognition," Proc. IEEE, vol. 86, no. 11, pp. 2278-2324, Nov. 1998.
[2]. A. Krizhevsky, I. Sutskever, and G. E. Hinton, "ImageNet classification with deep convolutional neural networks," in Advances in Neural Information Processing Systems, vol. 25, pp. 1097-1105, 2012.
[3]. Schober, P. and T. R. Vetter, "Logistic regression in medical research," Anesthesia & Analgesia, vol. 132, no. 2, pp. 365-366, 2021.
[4]. D. Mustafa Abdullah and A. . Mohsin Abdulazeez, “Machine Learning Applications based on SVM Classification A Review”, QAJ, vol. 1, no. 2, pp. 81–90, Apr. 2021.
[5]. A. N. Elmachtoub, J. C. N. Liang, and R. McNellis, "Decision trees for decision-making under the predict-then-optimize framework," in Proc. Int. Conf. Mach. Learn., PMLR, Nov. 2020, pp. 2858-2867.
[6]. F. I. Adiba, T. Islam, M. S. Kaiser, et al., "Effect of corpora on classification of fake news using naive Bayes classifier," Int. J. Autom. Artif. Intell. Mach. Learn., vol. 1, no. 1, pp. 80-92, 2020.
[7]. A. Esteva et al., "A guide to deep learning in healthcare," Nature Medicine, vol. 29, pp. 27-38, 2023.
[8]. S. Liu and X. Wang, "An improved method of medical images classification based on contrast learning," J. Jilin Univ. (Inf. Sci. Ed.), 2024.
[9]. M.E.H. Chowdhury, T. Rahman, A. Khandakar, R. Mazhar, M.A. Kadir, Z.B. Mahbub, K.R. Islam, M.S. Khan, A. Iqbal, N. Al-Emadi, M.B.I. Reaz, M. T. Islam, “Can AI help in screening Viral and COVID-19 pneumonia?” IEEE Access, Vol. 8, 2020, pp. 132665 - 132676.
[10]. Rahman, T., Khandakar, A., Qiblawey, Y., Tahir, A., Kiranyaz, S., Kashem, S.B.A., Islam, M.T., Maadeed, S.A., Zughaier, S.M., Khan, M.S. and Chowdhury, M.E., 2020. Exploring the Effect of Image Enhancement Techniques on COVID-19 Detection using Chest X-ray Images.
Cite this article
Sun,S. (2024). Pretrained-ResNet-based COVID-19 CT image classification. Theoretical and Natural Science,62,190-196.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Y. LeCun, L. Bottou, Y. Bengio, and P. Haffner, "Gradient-based learning applied to document recognition," Proc. IEEE, vol. 86, no. 11, pp. 2278-2324, Nov. 1998.
[2]. A. Krizhevsky, I. Sutskever, and G. E. Hinton, "ImageNet classification with deep convolutional neural networks," in Advances in Neural Information Processing Systems, vol. 25, pp. 1097-1105, 2012.
[3]. Schober, P. and T. R. Vetter, "Logistic regression in medical research," Anesthesia & Analgesia, vol. 132, no. 2, pp. 365-366, 2021.
[4]. D. Mustafa Abdullah and A. . Mohsin Abdulazeez, “Machine Learning Applications based on SVM Classification A Review”, QAJ, vol. 1, no. 2, pp. 81–90, Apr. 2021.
[5]. A. N. Elmachtoub, J. C. N. Liang, and R. McNellis, "Decision trees for decision-making under the predict-then-optimize framework," in Proc. Int. Conf. Mach. Learn., PMLR, Nov. 2020, pp. 2858-2867.
[6]. F. I. Adiba, T. Islam, M. S. Kaiser, et al., "Effect of corpora on classification of fake news using naive Bayes classifier," Int. J. Autom. Artif. Intell. Mach. Learn., vol. 1, no. 1, pp. 80-92, 2020.
[7]. A. Esteva et al., "A guide to deep learning in healthcare," Nature Medicine, vol. 29, pp. 27-38, 2023.
[8]. S. Liu and X. Wang, "An improved method of medical images classification based on contrast learning," J. Jilin Univ. (Inf. Sci. Ed.), 2024.
[9]. M.E.H. Chowdhury, T. Rahman, A. Khandakar, R. Mazhar, M.A. Kadir, Z.B. Mahbub, K.R. Islam, M.S. Khan, A. Iqbal, N. Al-Emadi, M.B.I. Reaz, M. T. Islam, “Can AI help in screening Viral and COVID-19 pneumonia?” IEEE Access, Vol. 8, 2020, pp. 132665 - 132676.
[10]. Rahman, T., Khandakar, A., Qiblawey, Y., Tahir, A., Kiranyaz, S., Kashem, S.B.A., Islam, M.T., Maadeed, S.A., Zughaier, S.M., Khan, M.S. and Chowdhury, M.E., 2020. Exploring the Effect of Image Enhancement Techniques on COVID-19 Detection using Chest X-ray Images.









