ccs concept: Applied computing, Life and medical sciences, Computational biology, Computational transcriptomics.
1. Introduction
Pancreatic ductal adenocancinoma is a lethal gastrointestinal cancer known for its early distant metastasis, extensive extracellular matrix deposition, disrupted vasculature, and immunosuppressive tumor microenvironment[1–10]. The immunosuppressive nature of the TME is marked by a high proportion of exhausted cytotoxic effector cells[11–13] and a population of myeloid cells, predominantly tumor-associated macrophages[14,15]. TAMs in primary and hepatic metastatic lesions arise from circulating peripheral monocytes and tissue-resident macrophages (TRM)[14–16]. Circulating monocytes are recruited by chemokines and differentiate into macrophages upon infiltration[14,17],while TRMs, derived from embryonic hematopoiesis, expand by in situ proliferation[16]. Macrophages exhibit both pro-tumor and anti-tumor functions[3,14–21]. In contrast to apoptotic cell debris phagocytosis, tumoricidal substances production, and cross-presentation that ignites adaptive immune response, TAMs generally contribute to tumor progression by facilitating tumor cell dissemination, early metastasis, immune evasion, and TME reprogramming, and by resisting chemotherapies[19,22–24] and immunotherapies[20,25–27]. However, macrophages can be externally stimulated to re-activate their tumoricidal abilities[21]. Due to the continuous gene profiles within TAM subtypes, strategies focus on modulating the pro-tumor to anti-tumor phenotype rather than direct depletion. β-glucans can reactivate Kupffer cells, liver-resident TRMs, to recognize and associate with seeding tumor cells, recruiting cytotoxic T cells, and shifting a subset of TAMs from pro- to anti-tumor properties[21].
The TME is highly dynamic and evolves with disease progression due to stage-dependent triggers[28–32]. Previous studies have described specific subtype TAM functions in isolation, either in primary[9,14,17,22,33] or hepatic metastatic[4,21,34] PDAC, but a comprehensive understanding of TAM correlations in these conditions still remains limited. Here, we reanalyzed monocytes/ macrophages (Mo/MΦ) from both primary and hepatic metastatic PDAC lesions. Identical subtypes show similar functions, and pseudotime trajectory and pseudobulk RNA analysis reveal shared metabolism reprogramming and inflammatory response between primary and hepatic metastatic TAMs. Single cell RNA sequencing and spatial transcriptomes (ST) show no exclusive distribution patterns of specific TAM subtypes in primary lesions, suggesting that phenotypic variations are primarily driven by cell-cell interactions (CCI) and intrinsic signaling rather than distribution variances. This highlights the importance of dissecting complex TME networks and factors that drive monocyte differentiation for therapeutic targeting and modulation of TAMs in PDAC.
2. Result
2.1. Subtypes of macrophages are relatively diverse across different conditions
To create a comprehensive transcriptional atlas of PDAC infiltrating TAMs, we reanalyzed scRNA-seq data 125422 cells of from primary PDAC and adjacent normal tissue, as well as 61916 cells of hepatic metastatic PDAC and normal tissue from patients and non-patients. We performed the analysis using single-cell variational inference (scVI)[55] after rigorous quality control. This process resulted in the identification of 20 cell types within primary (Figure 1a) and hepatic metastatic (Figure 1b) PDAC. Additionally, we further segregated 22877 cells of monocyte/macrophage (Mo/MΦ) population into eight subsets (Figure 1c, 1d, Supplementary Figure2): The CD14+/CD16+ monocytes and THBS1+ monocyte-like cell displayed characteristic monocyte marker patterns including CD14, FCER3A (CD16), Ficolin 1 (FCN1), S100 Calcium Binding Protein A8 and A9 (S100A8/S100A9). The phenotypes of the other four TAM subsets aligned with previous studies[17,56,57], SPP1+ TAM, IL1B+ TAM, and two tissue resident subsets, LYVE1+ FOLR2+ TAM, LYVE1- FOLR2+ TAM (Figure 1d).
We observed significant variation in the Mo/MΦ composition across different conditions (Figure 1e). Notably, the SPP1+ TAM and IL1B+ TAM populations were enriched in neoplastic lesions (P-value=0.00049 and 0.00915, respectively), while the tissue resident macrophage subset LYVE1+ FOLR2+ was enriched in non-neoplastic tissue (P-value=0.00359), suggesting a normal tissue resident macrophage phenotype in the liver and pancreas. Additionally, the landscape of tumor adjacent normal tissue Mo/MΦ differs quite from that in normal tissue from non-patients. The tumor-adjacent normal tissue Mo/MΦ landscape differed significantly from that in normal tissue from non-patients. The THBS1+ monocyte-like cell, a novel unreported metastatic PDAC TAM subtype, emerged as a unique subtype in hepatic metastases, prevailing over the other three conditions. Studies on this specific cell type in colorectal cancer[58–61] have suggested that THBS1+ monocyte-like cells are recruited from the bone marrow and contribute to the immunosuppressive microenvironment by inducing cytotoxic T cell exhaustion, thereby participating in metastasis. Therefore, the enrichment of THBS1+ monocyte-like cells in hepatic metastatic PDAC may also affect immune regulation, leading to inefficient adaptive immune cell infiltration—a critical aspect that merits further investigation into its pathological roles in the PDAC metastatic context.
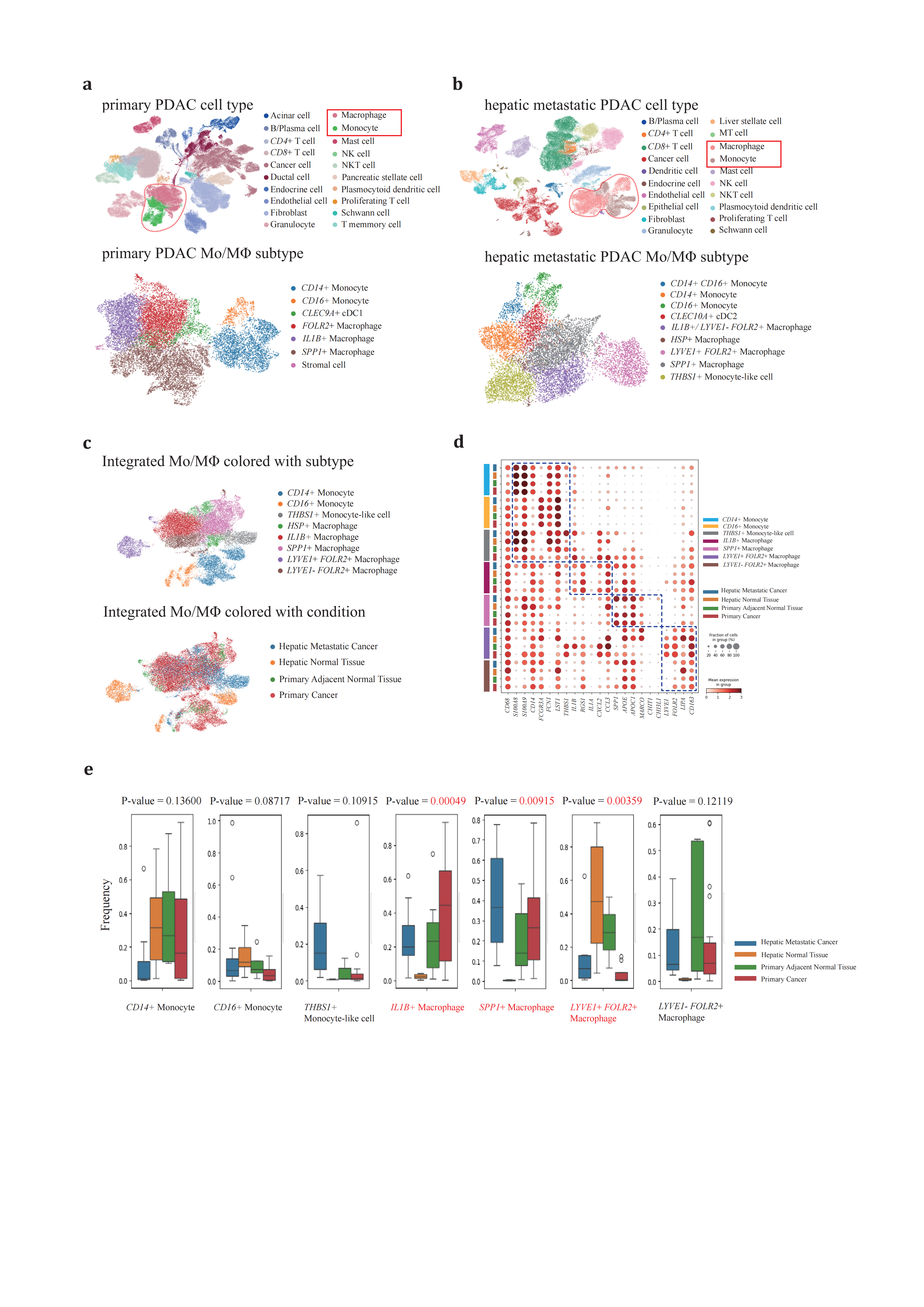
Figure 1. Subtypes of macrophage are conservative across different conditions(a-b), clustering of (a) primary PDAC and adjacent normal tissues and (b) hepatic metastatic PDAC and normal tissues. (c), UMAP projection of Mo/MΦ from primary/adjacent normal tissues and hepatic metastatic/normal tissue PDACs, colored according to graph-based clustering (up panel) or sample origin (down panel). (d), Dot plot depicting expression information of representative Mo/MΦ marker genes. (e), percentage of each Mo/MΦ subtype across four conditions. ANOVA method is used to measure specific subtype variance. Samples reanalyzed in this study are generalized in supplementary table1.
These findings underscore the considerable diversity in Mo/MΦ composition across various conditions, reflecting their distinct physiological roles. Specifically, the SPP1+ TAM and IL1B+ TAM populations were predominantly localized within neoplastic lesions and adjacent normal tissues, highlighting their significant involvement in tumor dynamics.
2.2. Two developmental trajectories of peripheral blood monocyte derived IL1B+ and SPP1+ TAMs
To elucidate the developmental dynamics of peripheral blood monocytes that are recruited and infiltrate neoplastic tissues, we conducted pseudotime trajectory analysis using Monocle2[62] aiming to infer the monocyte cell fates under these conditions (Figure 2a). The Mo/MΦ were organized along the trajectory, with each cell identity properly projected onto the trajectory plot. The CD14+/CD16+ monocytes predominantly occupied State 9 and served as the progenitor cells for tumor-infiltrating macrophages (Figure 2b, 2c), an observation that was also corroborated by the high density of monocytes in hepatic normal tissue.
From the branch point 4, differentiating cells underwent bifurcating cell fate inductions (Figure 2d). One branch terminated at State 3, while the other clusters continued in the opposite direction through State 4 and 5, ultimately stopping at State 7 and 8. This bifurcation pattern prompted further investigation into the cell types distributed along these two distinct paths from branch point 4. State 3 was primarily composed of IL1B+ TAMs, whereas SPP1+ TAMs spanned from State 4 to 8 (Figure 2e). Notably, State 7 was shared nearly equally by two cell types, suggesting an ‘intermediate’ phenotype between IL1B+ and SPP1+ TAMs in high resolution. This intermediate state potentially represents a phenotypic transition between TAMs.
Moving forward from branch point 4, we profiled the dynamic regulations of gene expression (Figure 2f, 2g). State 3 cells revealed upregulated genes involved in the cell cycle checkpoint and a simultaneous upsurge in pro-inflammatory genes through several pathways, resembling the traditional ‘M1-like’ macrophage phenotype. Early Growth Response 1 (EGR1), a crucial transcriptional factor for monopoiesis, was activated through the EGFR/RAS/MEK/ERK pathway and bound to a large set of inflammatory enhancers, thereby upregulating the expression of inflammatory-related genes[63–65]. Kruppel-like transcription factor-6 (KLF6) promoted pro-inflammatory gene expression through the enhancement of nuclear factor κB (NFκB) signaling pathway[66–68]. Cytokines and chemokines are families of secreted proteins that function in inflammatory and immunoregulatory processes. Cytokines and chemokines, families of secreted proteins involved in inflammatory and immunoregulatory processes, were expressed in State 3 cells, including IL1B, CCL3, CCL4, CCL4L2, and CXCL2, which recruit regulatory T cells and neutrophils, exacerbating local inflammation and immunosuppression[17,69–71]. Interestingly, cells in State 3 retained antigen-peptide presenting ability by highly expressing major histocompatibility complex (MHC) subunits and associated proteins, CD74, CD83. HLA-DRB5, HLA-DPA1, HLA-DPB1, HLA-DRA, HLA-DRB1, HLA-DQA1, which can partially compensate for the absence of dendritic cells in PDAC[1,2].
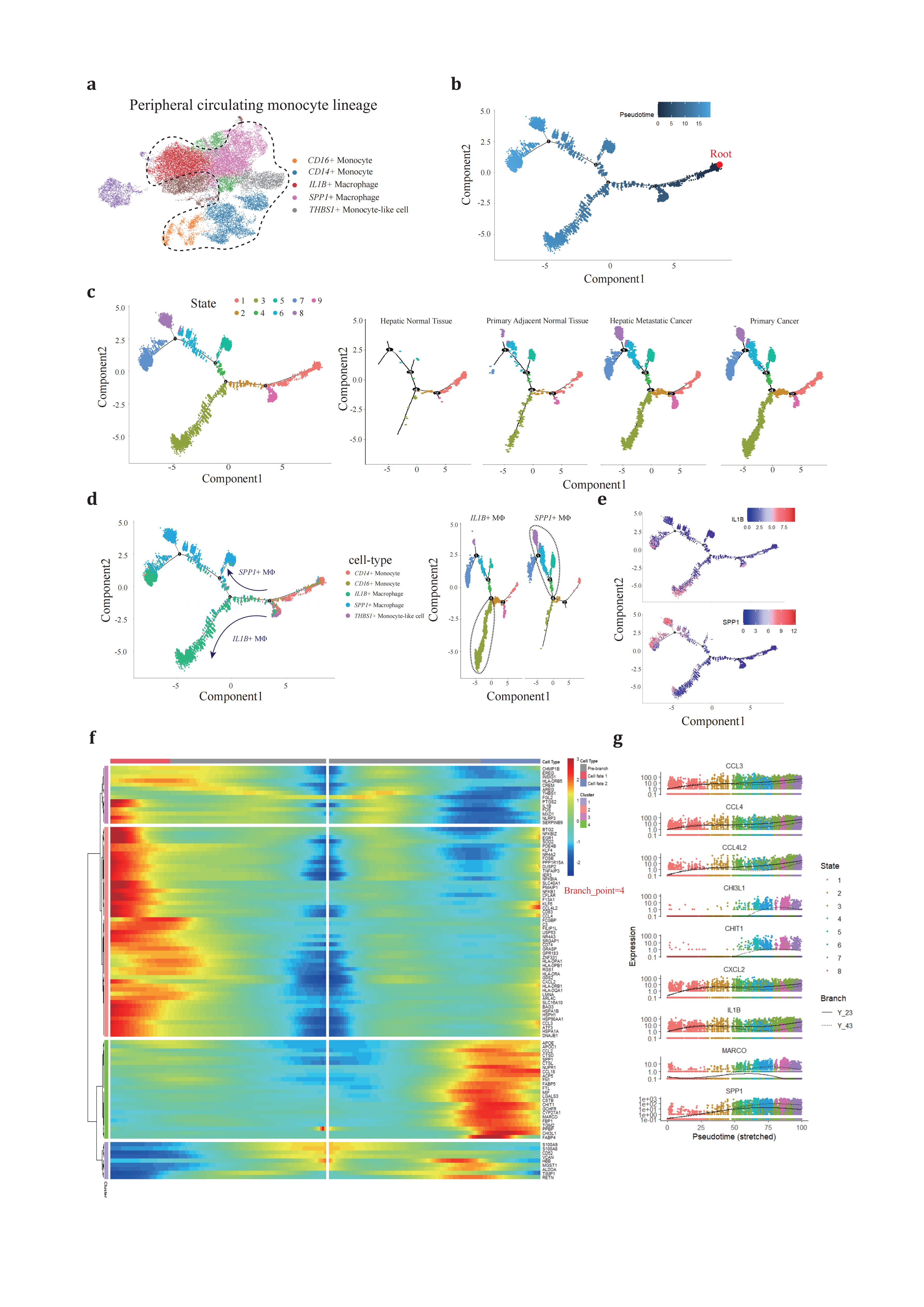
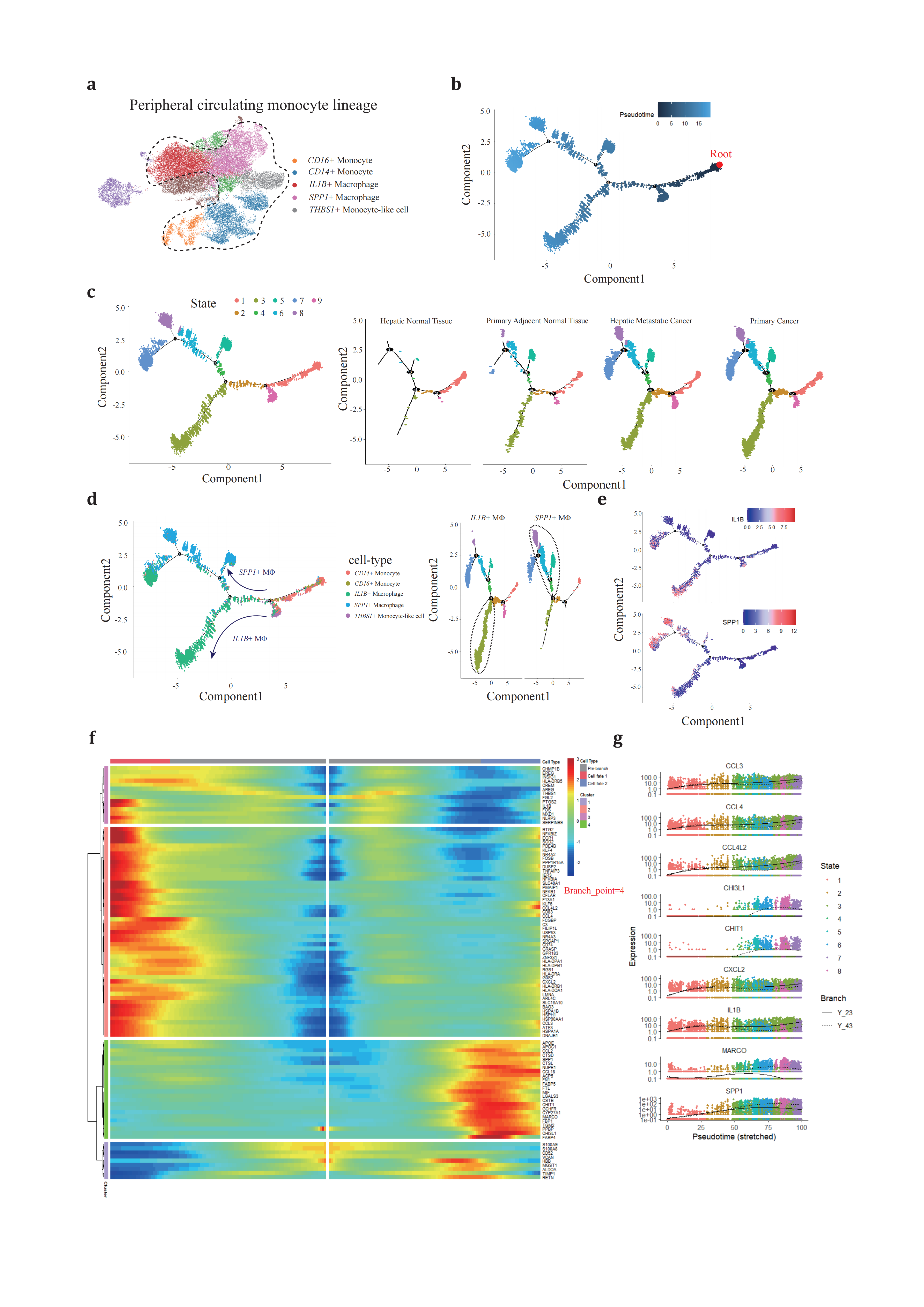
Figure 2. Different development trajectory of IL1B+ and SPP1+ peripheral macrophage(a), peripheral blood original Mo/MΦ populations are selected to reconstruct pseudotime trajectory. (b), pseudotime trajectory is reconstructed on the selected populations containing one branch point. Branch occupied by monocytes is chosen as root referring to figure2c and figure2d. (c), cells of different states are denoted with different colors (left panel) across conditions (right panels). (d), each Mo/MΦ subtype (left panel) is projected onto the trajectory plots with different colors. Cell states are denoted with two specific subtypes, IL1B+ and SPP1+ TAMs, with the most dominant branch circled. (e), IL1B and SPP1 expression along trajectory plots. (f), heatmap presents a differentially expressed gene profile from branch point 4, each row represents expression level of each gene along the two branch trajectories. (g), pseudotime kinetics of representative genes from the branch point 4 of the trajectory to cell fate 1 (solid line) or cell fate 2 (dashed line), with each dot representing a single cell and color-coded by cell state.
Conversely, the SPP1+ TAM following the opposite route exhibited activation of extracellular matrix (ECM) remodeling(TIMP1[72], SPP1[73], CSTB, CSTD and CSTL[74,75]), scavenger receptor mediated phagocytosis (MARCO[76]) and involved in metabolism and recycling like lipoprotein metabolism (APOE and APOC1[77]). Compared to cells in State 3, these cells exhibited less interaction with other immune components.
These findings confirm the existence of dual directional developmental trajectories for peripheral blood monocytes, each characterized by specific stimuli that lead to unique cellular fates: one specialized in ECM remodeling and the other in contributing to the inflammatory response.
2.3. Similar pathological transition of TRMs from normal tissue to neoplastic lesion
Based on previous reports that LYVE1- FOLR2+ TRMs exhibit fetal-liver macrophage characteristics and contribute to onco-fetal reprogramming in hepatocellular carcinoma (HCC)[78], we hypothesized that LYVE1+ FOLR2+ TRMs might serve as a ‘reserve’ for the in-situ proliferation and functional transition of LYVE1- FOLR2+ TRMs. To test this hypothesis, we expanded our analysis to include both TAM subsets across four conditions (Figure 3a). We employed the same trajectory inference strategy and chose the LYVE1+ FOLR2+ macrophage state (State 2) as the root of our analysis due to its predominant presence in hepatic normal tissue (Figure 3b).
Through pseudotime trajectory analysis, TRMs were ordered along the trajectory, consisting of one root (State 2) and two termini corresponding to two distinct cell fates (State 1 and 3) (Figure 3c). TRMs positioned along State 2, particularly those at the termini, underwent continuous reprogramming (Figure 3d). We proposed that cells differentiating out of State 2 (diverging from branch point 1) resembled the populations of peripheral blood monocytes at branch point 4 and represented a critical transitional phase. Cells in State 1 showed strong correlations with SPP1+ TAM (ALDOA, LPL, APOE, APOC1, CSTB, CTSD, TSPO, CHIT1, and CHI3L1), while those in State 2 exhibited phenotypic similarities to IL1B+ TAM (IL1B, KLF6, RGS1, and RGS2) (Figure 3e-g).
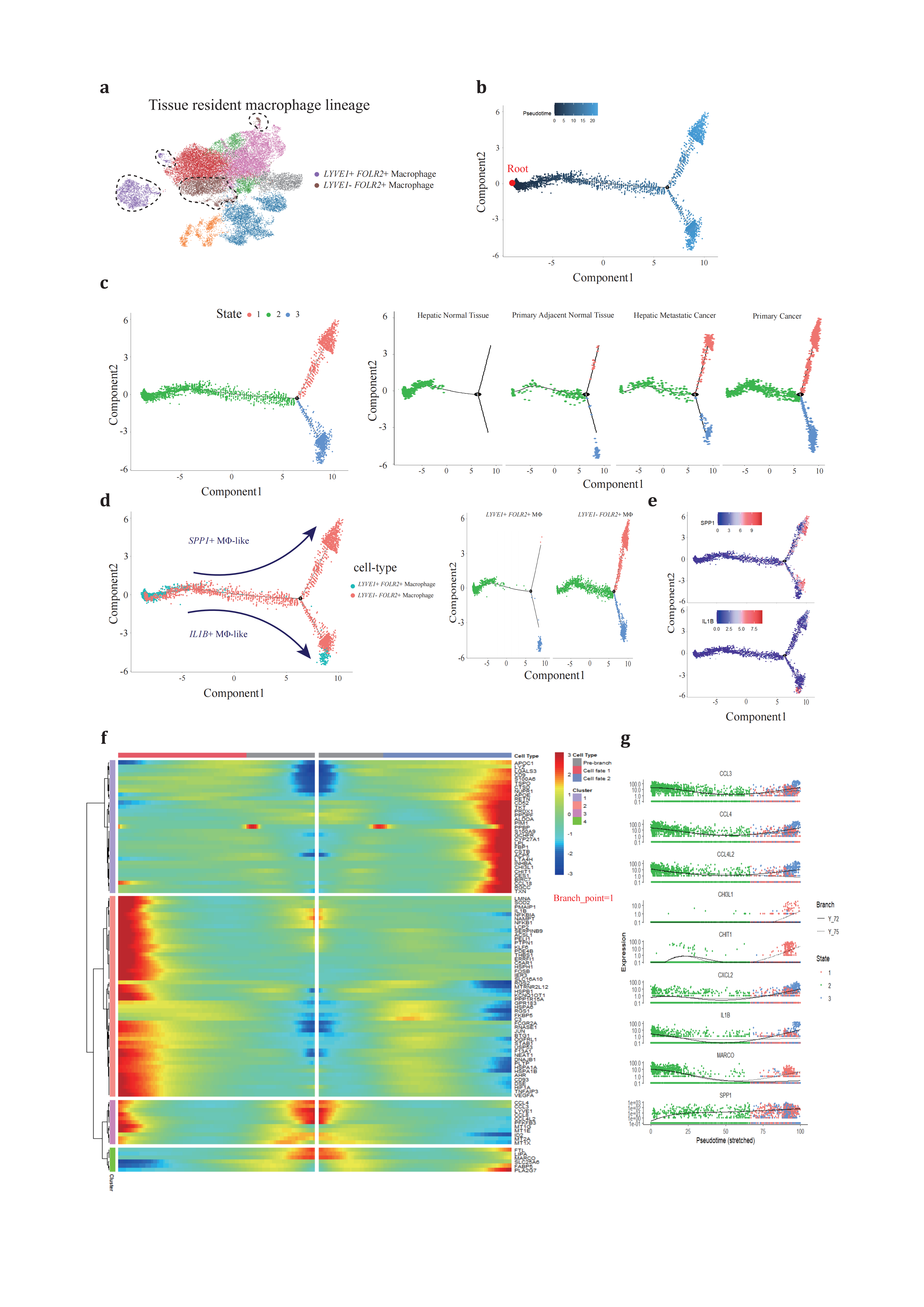
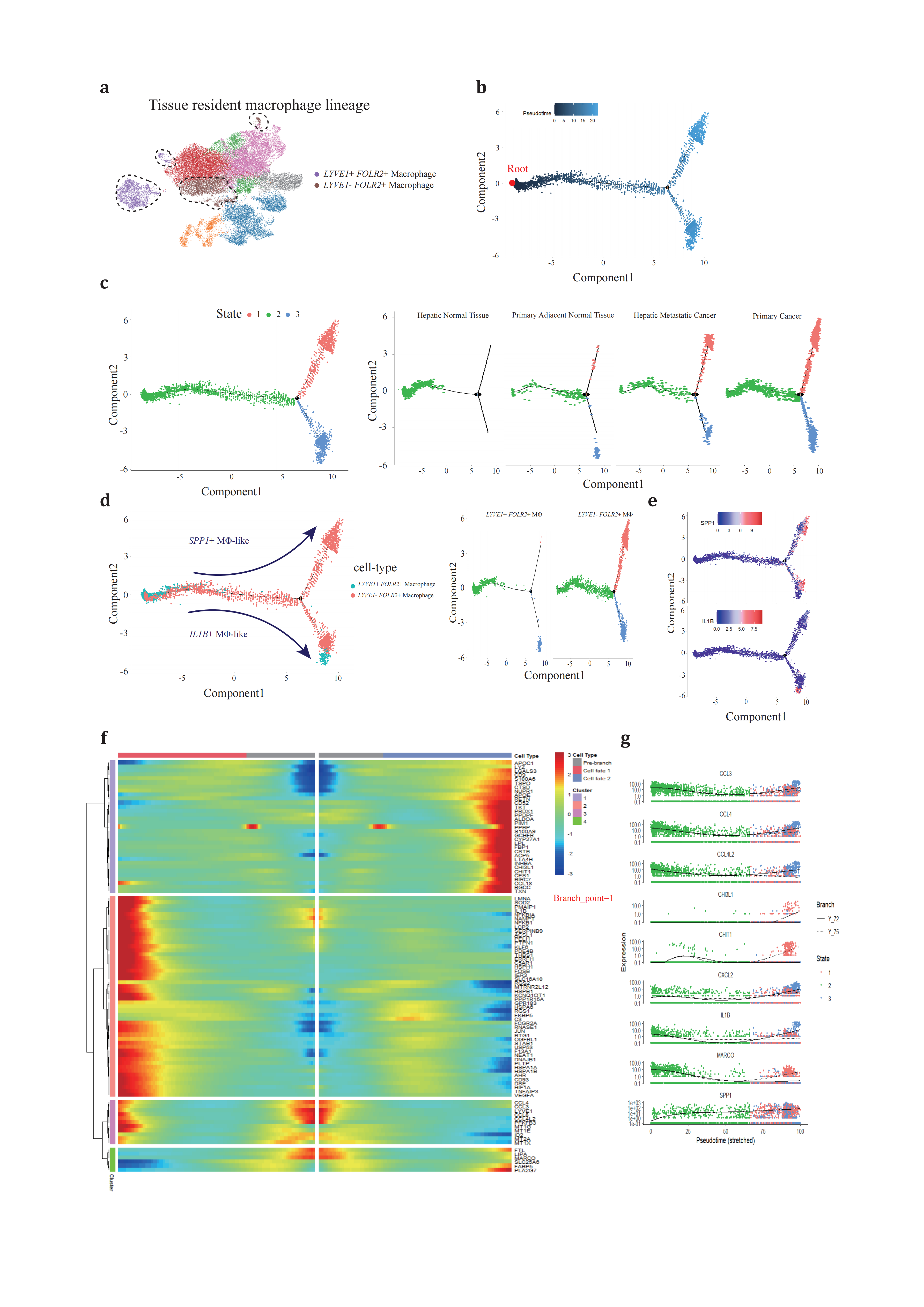
Figure 3. Different development trajectory of TRMs across conditions(a), TRMs are selected to reconstruct pseudotime trajectory. (b), pseudotime trajectory is reconstructed on the selected populations containing one branch point. Branch occupied by LYVE1+ FOLR2+ TRM is chosen as root. (c), cells of different states are denoted with different colors (left panel) across conditions (right panels). (d), two TRM subtypes (left panel) are projected onto the trajectory plots with different colors. (e), representative marker genes projected onto trajectory plots. (f), heatmap presents a differentially expressed gene profile from branch point, each row represents expression level of each gene along the two branch trajectories. (g), pseudotime kinetics of representative genes from the branch point of the trajectory to cell fate 1 (solid line) or cell fate 2 (dashed line), with each dot representing a single cell and color-coded by cell state.
These findings suggest that SPP1+ TAMs and IL1B+ TAMs, despite their different origins, may represent two functional paradigms of TAMs, fulfilling distinct roles in either pro-inflammation or extracellular matrix remodeling and metabolism. The default clustering and trajectory projection indicate that these cells converge toward identical fates, emphasizing the importance of further studies to investigate the determinants that trigger divergence in TAM cell fates. Such investigations could significantly advance our understanding of TAM functional heterogeneity.
2.4. MΦ polarization across conditions reveal distinctive signaling network prospective
MΦ polarization refers to the differentiation of macrophages towards specific phenotypes[79]. The traditional concept of MΦ polarization is overly simplified, categorizing macrophages into either pro-inflammatory M1 or anti-inflammatory M2 types, which correspond to anti-tumor and pro-tumor effects in the TME, respectively. However, high-resolution technologies such as scRNA-seq and ST have begun to challenge this binary classification by providing detailed profiles of individual cell transcriptomes and their spatial organization within immunosuppressive niches, casting doubt on the reliability of a strict M1/M2 dichotomy. To characterize the functional skewing of specific subtypes across conditions, we conducted pseudobulk RNA analysis to identify DEGs and perform functional enrichment analysis.
Functional enrichment based on The Molecular Signatures Database (MSigDB) hallmark revealed significant functional variance across conditions and subtypes (Figure 4a). IL1B+ and SPP1+ TAMs exhibited polarized gene profiles (Figure 4b). The IL1B+ TAM revealed the most significant DEGs profiles in primary PDAC and hepatic metastatic PDAC (Figure 5b) as well as in primary adjacent normal tissue and primary PDAC (Figure 5c), indicating a co-evolution with the progressive TME. Consistent with pseudotime trajectory analysis results, the IL1B+ TAM is posited to be the source of local inflammation response, with elevated MTORC1 signaling and IL2/STAT5 signaling, along with TGFβ signaling and canonical TNFɑ signaling via the NFKB pathway hyperactivation (Figure 4c-4e). This trend is observed in both primary PDAC and hepatic metastatic PDAC progression.
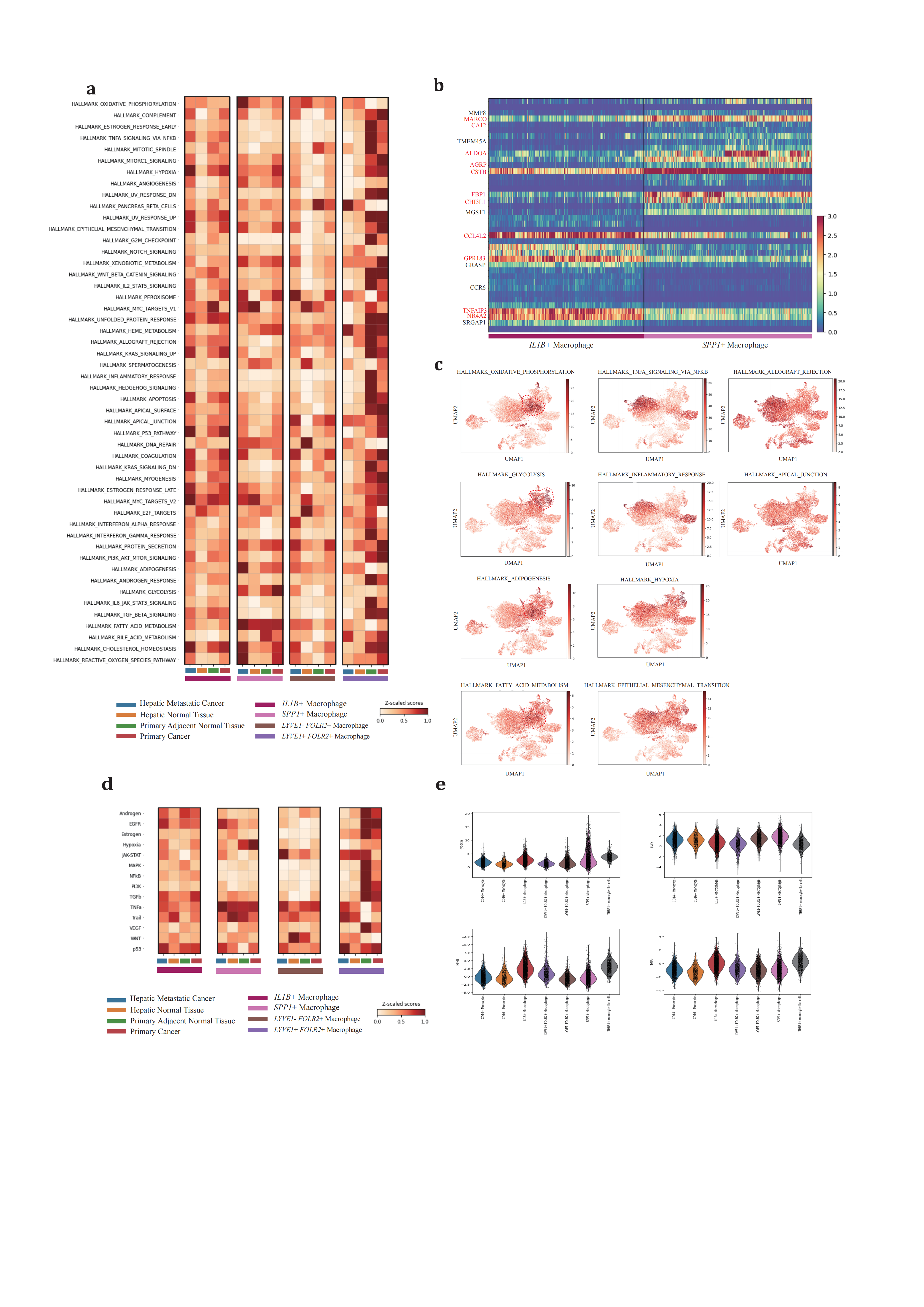
Figure 4. Differentially regulated pathway and functional enrichment in TAMs(a), heatmap showing normalized functional enrichment scores of four cell subtypes across four different conditions. (b), differentially expressing genes between IL1B+ and SPP1+ macrophage by scVI. (c), UMAP projection of normalized functional enrichment scores for representative cancer-related hallmarks. (d), heatmap showing normalized pathway activity scores of four cell subtypes across four different conditions. (e), violin plots visualize relative activation degrees. This analysis utilizes hallmark gene sets from The Molecular Signatures Database (MSigDB). Over-representation analysis (ORA) is conducted with default parameters to obtain functional enrichment scores for each cell subtype.
For the SPP1+ subtype, there were no significant DEGs in primary adjacent normal tissue/primary PDAC (KLK1, AMY2B) and primary PDAC/hepatic metastatic PDAC (PSME1, TIMP1, GPNMB, APOC2, EEF1G, RPL17) (Figure 5a). GPNMB, a transmembrane glycoprotein overexpressed in various cancers, has been shown to be an essential mediator in epithelial-to-mesenchymal transition in glioblastoma[80]. PSME1, which is involved in immunoproteasome assembly and efficient antigen processing, positively correlates with anti-tumor properties in gastric cancer by enhancing the infiltration of cytotoxic immune cells[81]. The upregulation of PSME1 in hepatic metastatic PDAC aligns with an initial effective immune response. TIMP1, a natural inhibitor of matrix metalloproteinases, has been demonstrated to directly trigger the formation of neutrophil extracellular traps in primary human neutrophils, which is dependent on the interaction of TIMP1 with its receptor CD63 and subsequent ERK signaling in PDAC[72,82]. In contrast to PSME1, TIMP1 contributes to the early metastatic TME construction.
ECM remodeling and metabolic reprogramming are considered dominant altered characteristics of the SPP1+ TAM (Figure 4a, 4c-e). In response to deficient oxygen and nutrient supply in the TME, the SPP1+ TAM adapts by relying on oxidative phosphorylation, glycolysis, fatty acid metabolism, and heme metabolism to varying degrees across conditions, promoting the production of reactive oxygen species. Compared to primary PDAC, hypoxia and nutrient deficiency are alleviated in hepatic metastatic neoplasms, supporting adipogenesis and initiating metastatic development. The SPP1+ TAM also engages in the phagocytosis and recycling of ECM deposited components to maintain self-survival. In this way, by shaping the ECM stromal composition, it enhances tumor cell mobility and facilitates metastasis.
The LYVE1- FOLR2+ TRMs also undergo a similar educational process as the IL1B+ or SPP1+ TAMs. Therefore, it is rational for this subtype to exhibit no predominant DEGs change and pathways activation across primary PDAC and hepatic metastatic PDAC during carcinogenesis (Figure 5d).
These comprehensive findings underscore the complexity of MΦ polarization and its evolution alongside tumor progression, emphasizing how chronic inflammation, worsened hypoxia, and nutrient deprivation drive phenotypic shifts in MΦ populations.
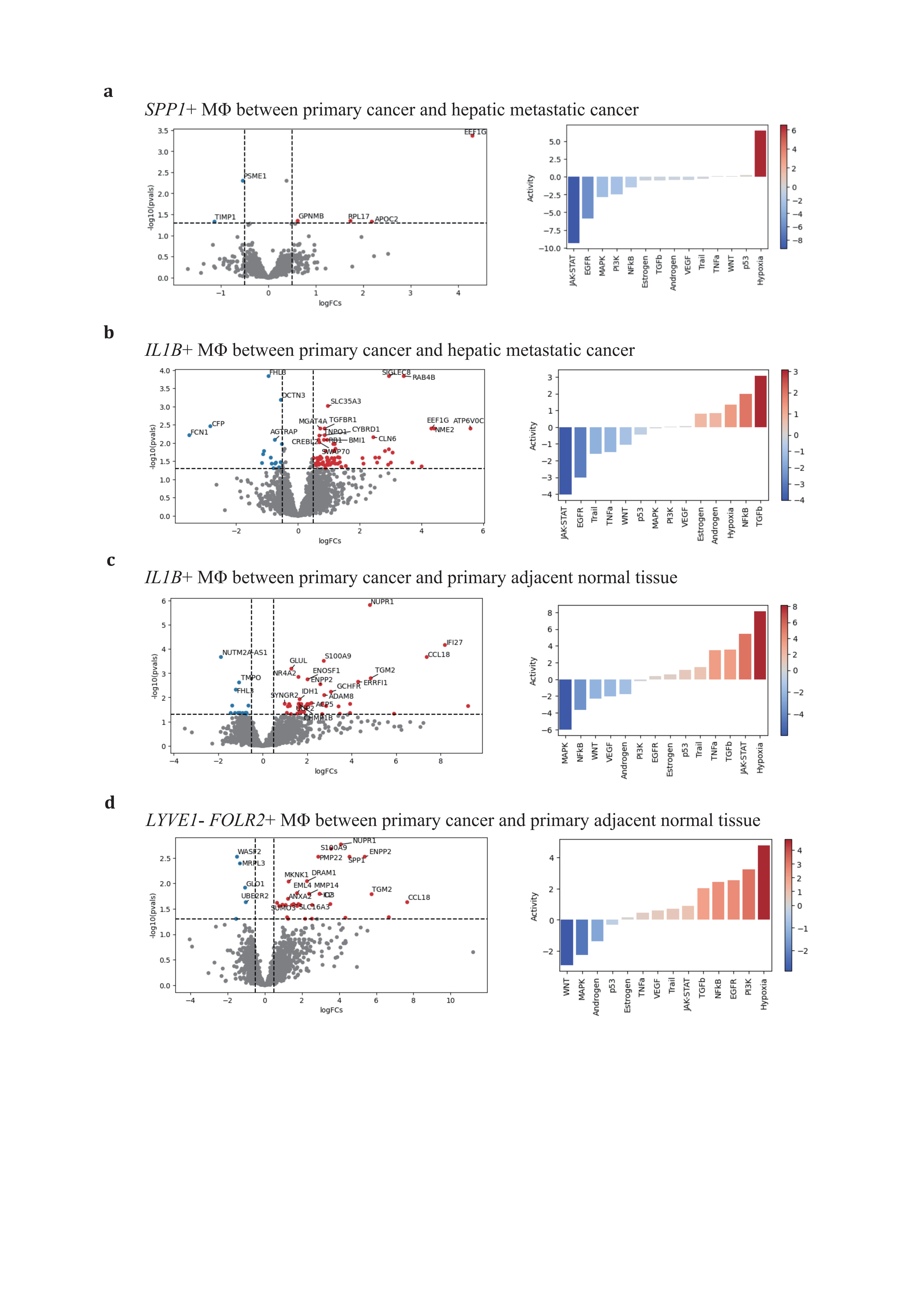
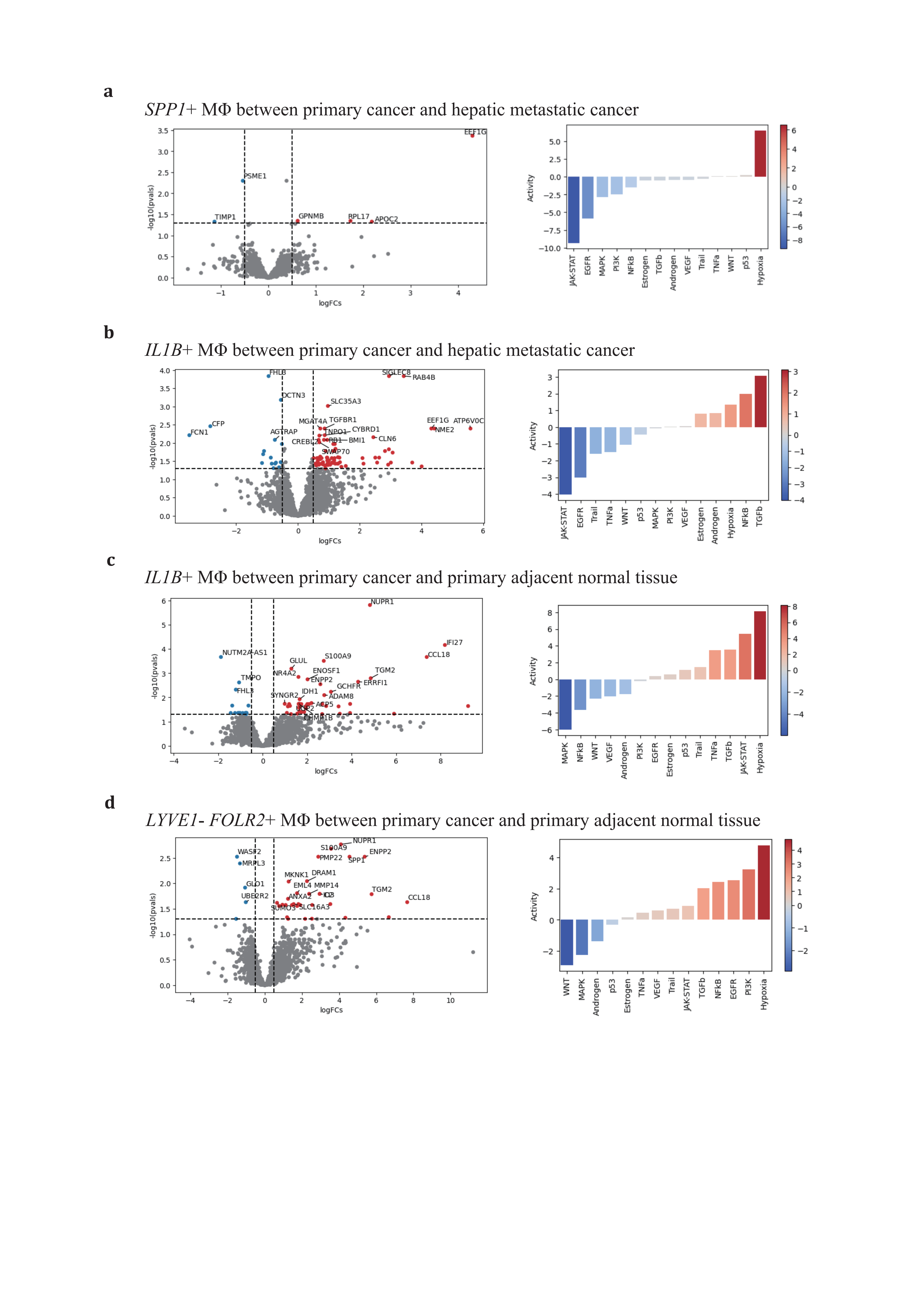
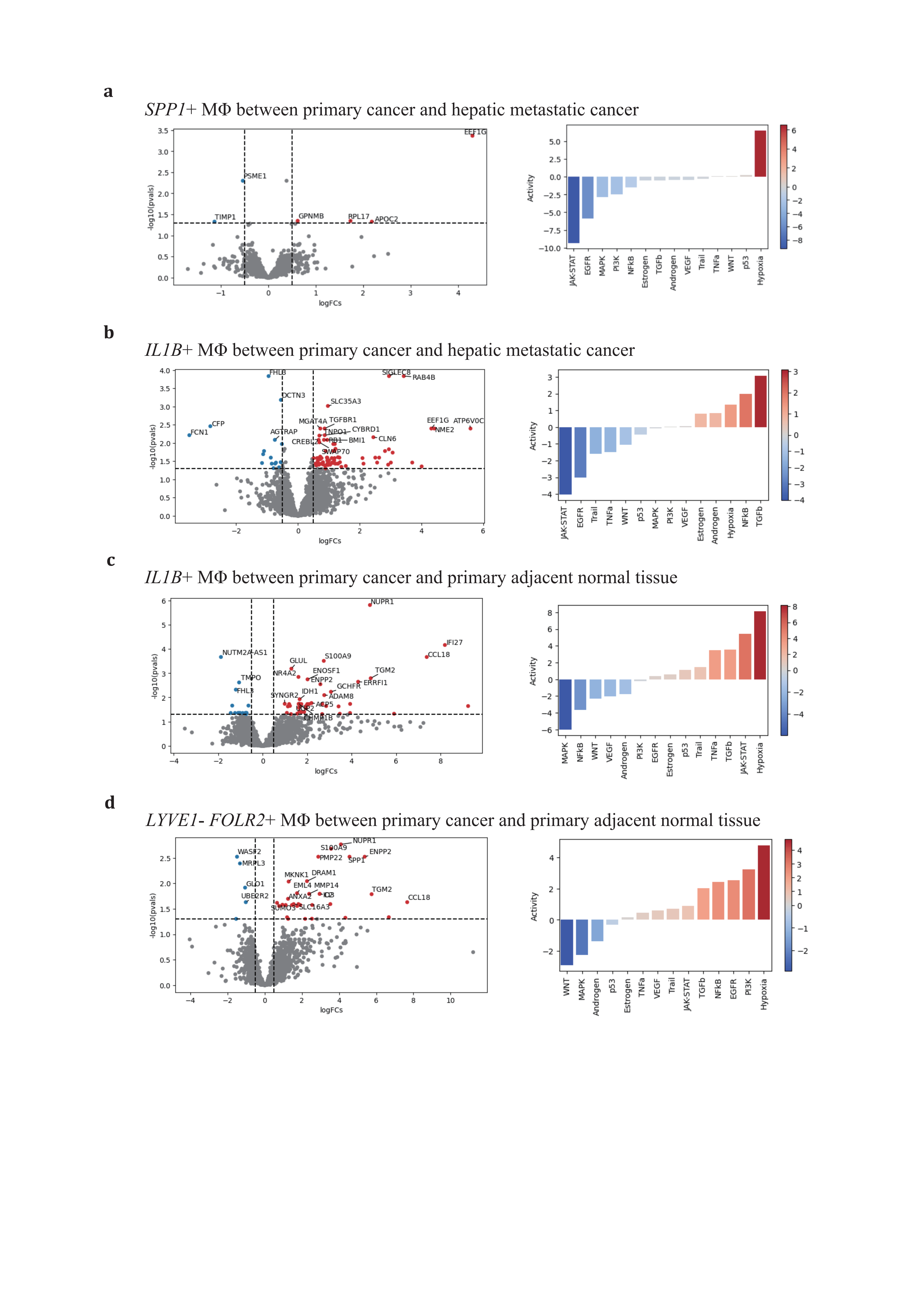
Figure 5. Distinctive expression profiling of SPP1+, IL1B+, and LYVE1- FOLR2+ TAMs(a-d), volcano plots of 3 major TAMs depict adaptive differentially expressing gene profiles across conditions (left panel), and corresponding activated gene pathways (right panel). (a), SPP1+ TAM in primary vs hepatic metastatic; (b), IL1B+ TAMs in primary vs hepatic metastatic and (c) primary cancer vs primary adjacent normal tissue; (d), LYVE1- FOLR2+ TAMs in primary cancer vs primary adjacent normal tissue.
2.5. Intertwined networks cause MΦ polarization but not distribution variance
The spatial organization of cell populations is crucial for the formation of cell-cell interaction (CCI) networks that shape cell relationships and phenotypes[83]. Cells that are co-localized have the opportunity for direct ligand-receptor interactions, while cells that are dispersed must communicate remotely through secreted cytokines and the ECM. A study highlighting the synergistic promotion of tumor progression by SPP1+ TAMs and FAP+ fibroblasts suggests that the spatial proximity of these cell populations is key to their coordinated signaling and the limitation of immune cell infiltration into the tumor core[84].
Inspired by this concept, we sought to investigate whether macrophage polarization is related to their distribution within the tumor microenvironment. We integrated primary PDAC ST and scRNA-seq data using GraphST[85] (Figure 6a,b). to analyze the spatial organization of cell populations. We found that TAMs are largely co-localized without distinctive distributive patterns, suggesting uniform distribution of nutrients and oxygen among them. The presumed progenitor cell, CD14+ monocytes, partially overlaps with other TAMs. We hypothesized that regulatory functions are most persistent. Regarding TAMs, our findings indicate that these macrophages are almost entirely co-localized at the same locations without discernible distinct distribution patterns, which excludes the possibility that metabolites specifically contribute single subtype development. However, the metabolic capacities measured by METAFlux[86] are disparately different. Notably (Figure 6c), SPP1+ TAMs, in particular, exhibit a high uptake of glucose and release of lactate, indicating a preference for anaerobic glycolysis.
The observed dual directional polarization of macrophages, with IL1B+like pro-inflammatory and SPP1+like metabolism reprogramming, is attributed to both extrinsic signaling triggers and intrinsic signaling orchestration, which together form a feedback loop. CCI networks visualized through ligand-receptor pairs interactive strength suggest that these interactions play a pivotal role in regulating physical interactions with other cell members and are key to immune response, adhesion, and migration processes in macrophage populations. These results suggest that physiological distribution plays a minimal role in macrophage polarization, with the intertwined regulatory networks of CCI synergistically educating differentiating macrophages. However, the question remains whether differentiating macrophage populations are driven to interact with specific cell populations compulsorily or if these interactions follow a more stochastic process. To better understand the dynamic regulatory mechanisms governing macrophage differentiation and function in the TME, real-time monitoring of cell behavior and interactions is essential. Such an approach could provide deeper insights into the complex interplay between cell types and offer potential strategies for therapeutic intervention.
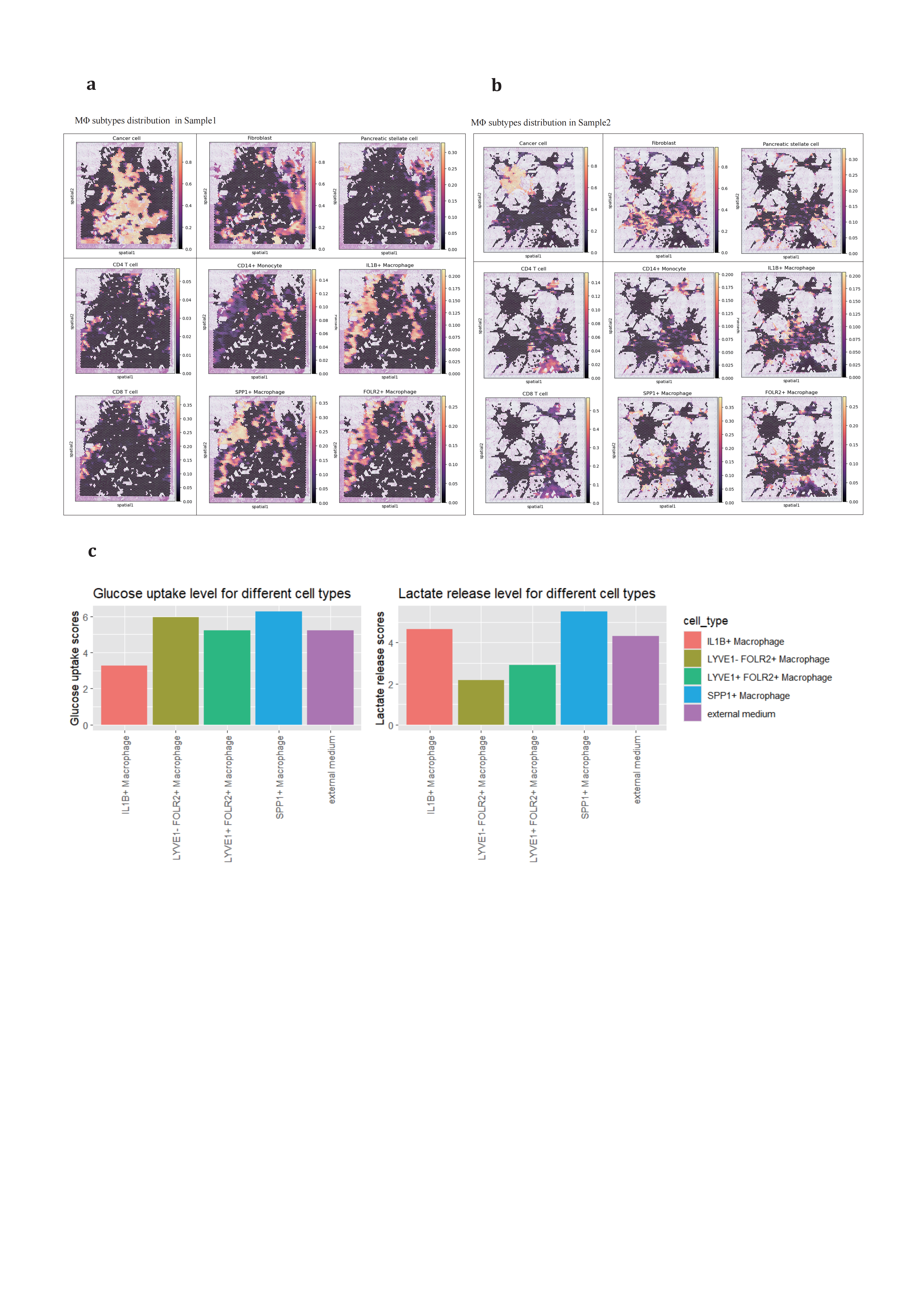
Figure 6. Spatial organization and metabolic ability measure of TAMs populations in PDAC(a-b), integrating scRNA-seq and ST showing spatial organizational patterns of major stromal, adaptive immune and Mo/MΦ populations in primary PDAC sample slice, (a) in sample SS1905133_processed and (b) in sample SS1923404_processed. (c), METAFlux measuring metabolic ability of glucose and lactate among cell subtypes. The normalized MRAS was calculated using the GPR approach and corresponding fraction parameters are assigned as followed: IL1B+ TAM (0.3), LYVE1- FOLR2+ TAM (0.2), LYVE1+ FOLR2+ TAM (0.1), and SPP1+ TAM (0.4). These parameters were used to calculate the metabolic flux for each cell type.
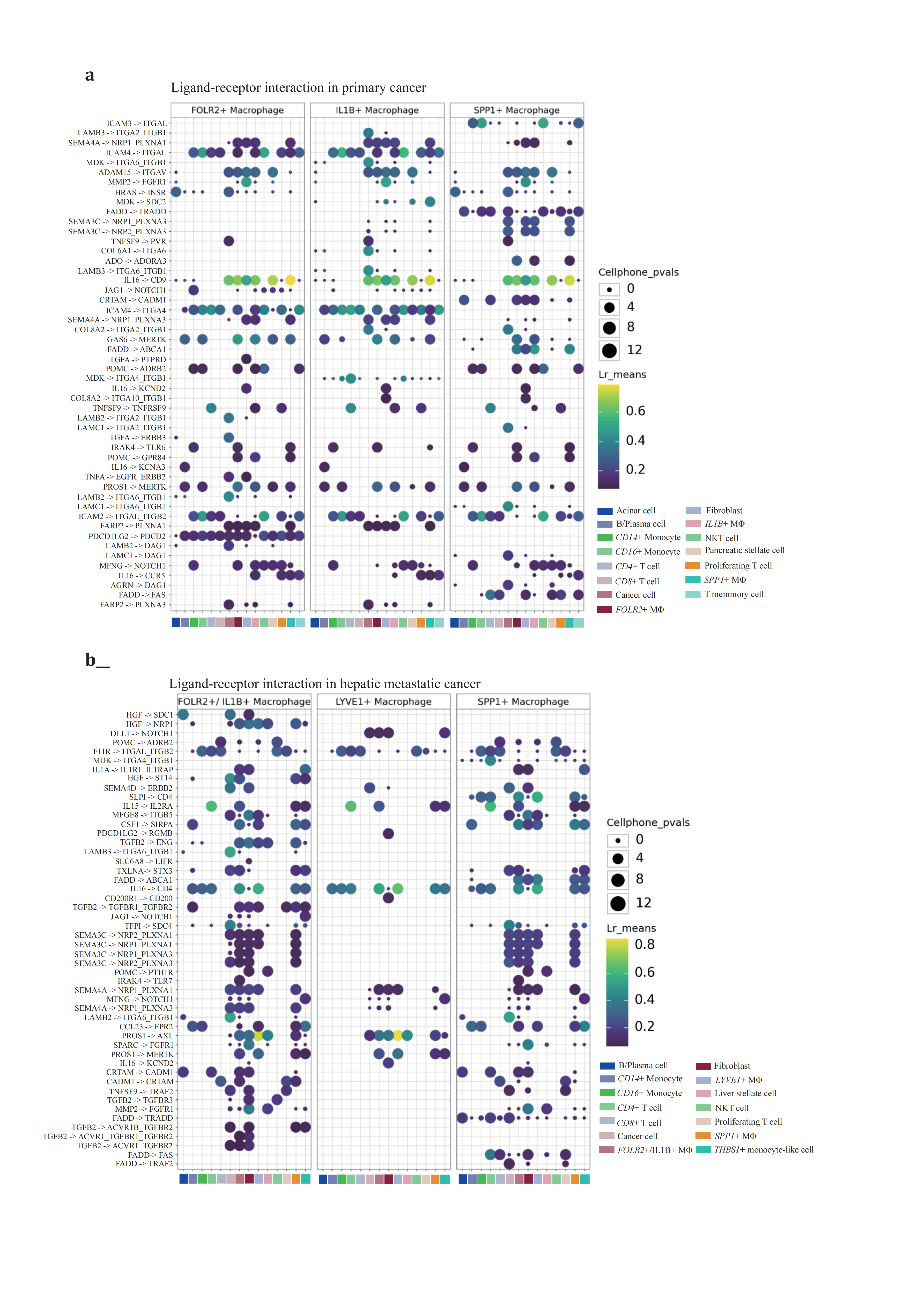
Figure 7. LR inference of TAMs with interactive cell populations(a-b), LR inference of TAMs with selected adjacent cell populations based on spatial inference in (a) primary PDAC and (b) hepatic metastatic PDAC. CellPhoneDB method LIANA offered by default parameters. Top 50 genes were used to visualize the interactive networks of TAMs and other cells in both primary and hepatic metastatic lesions.
3. Discussion
Current comprehension of PDAC TME progresses a lot due to upgraded sequencing technologies. As mentioned, a number of studies have uncovered the heterogeneous landscape of myeloid cell populations in isolated conditions[4,14,17,21,22,33,34,47]. Our study extends this kind of research by reanalyzing monocytes/macrophages collected from both primary and metastatic lesions. Through both horizontal and vertical comparisons, we have delineated the main developmental trajectories of monocytes—from recruitment to infiltration and differentiation—into two predominant paradigms: the IL1B+ like TAMs, which drive pro-inflammatory responses, and the SPP1+like TAMs, which contribute to extracellular matrix remodeling and metabolic reprogramming. Irrespective of their final cell fates, both can promote EMT and immunosuppressive niche construction. In our analysis, we noticed that overlap between SPP1+ TAM and IL1B+ TAM exists. This kind of “double positive” cell populations suggests a transition between subtypes continues even in terminal states, which implies a strong plasticity among macrophages during cancer development[87,88]. Interestingly, our findings highlight that the overall progression and impact of TAMs within the TME are profoundly influenced by extrinsic CCI networks. Underdeveloped CCI networks have been observed to partially alleviate conditions such as hypoxia, release of immunosuppressive cytokines, suggesting potential combined therapeutic targets.
Looking ahead, the attractive field for future exploiting, we supposed, is to figure out the cell fate decision factors to the full extent of hindering TAMs participating in cancer progression and drug resistance acquirement. While directly targeting the KRAS mutation in tumor cells represents a straightforward therapeutic approach with the potential to eliminate neoplastic cells, this strategy does not address the issue of disseminated tumor cells that can relocate and form new neoplasms. TAMs play a critical role in these metastatic scenarios, facilitating initial tumor seeding and the development of an early immunosuppressive niche that supports tumor growth and immune evasion. Identifying and targeting the factors that dictate TAM polarization and function can disrupt the supportive role TAMs play in tumor survival and metastasis[26,89–91]. This could involve inhibiting the signaling pathways that lead to the recruitment and pro-tumor activation of TAMs or promoting pathways that drive their anti-tumor activities.
One therapeutic strategy could be to hinder the immunosuppressive functions of TAMs while simultaneously restoring their antigen-presenting capabilities[92–94]. This dual approach has the potential to reactivate the adaptive immune response, which is often suppressed in PDAC. By re-engaging cytotoxic T cells and other components of the adaptive immune system, it may be possible to enhance the efficacy of immunotherapies, including immune checkpoint inhibitor like PD1/PD-L1 pair, which has shown limited success in PDAC due to the dense immunosuppressive TME.
Additionally, combining TAM modulation with other treatments, such as dissecting dense extracellular matrix that allows tumoricidal agents penetration and strategies towards vasculature recovering that continuously supplies nutrient and oxygen for infiltrating adaptive immune cell normal functions could further improve therapeutic outcomes[1]. The use of agents that can reprogram TAMs to support anti-tumor immunity, such as toll-like receptor agonists may also provide a synergistic effect with these treatments.
By fully understanding and manipulating the factors that govern TAM cell fate, we assume this can open a new therapeutic window in the clinical treatment of PDAC, transforming the management of this aggressive disease and improving patient survival rates.
Acknowledgements
I hereby extend my sincere gratitude to Dr. Guangwei Zhang of the University of Southern California and Dr. Bo Liu of the Memorial Sloan Kettering Cancer Center for their theoretical guidance in this research. Additionally, I express my heartfelt thanks to Mr. Mark Sanborn of the Viral Diseases Branch at the Walter Reed Army Institute of Research in Silver Spring, Maryland, USA, for his technical support.
References
[1]. Halbrook CJ, Lyssiotis CA, Pasca Di Magliano M, Maitra A. Pancreatic cancer: Advances and challenges. Cell. 2023;186(8):1729-1754. doi:10.1016/j.cell.2023.02.014
[2]. Luo W, Wen T, Qu X. Tumor immune microenvironment-based therapies in pancreatic ductal adenocarcinoma: time to update the concept. J Exp Clin Cancer Res. 2024;43(1):8. doi:10.1186/s13046-023-02935-3
[3]. Joseph AM, Al Aiyan A, Al-Ramadi B, Singh SK, Kishore U. Innate and adaptive immune-directed tumour microenvironment in pancreatic ductal adenocarcinoma. Front Immunol. 2024;15:1323198. doi:10.3389/fimmu.2024.1323198
[4]. Zhang S, Fang W, Zhou S, et al. Single cell transcriptomic analyses implicate an immunosuppressive tumor microenvironment in pancreatic cancer liver metastasis. Nat Commun. 2023;14(1):5123. doi:10.1038/s41467-023-40727-7
[5]. Bulle A, Lim KH. Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Sig Transduct Target Ther. 2020;5(1):249. doi:10.1038/s41392-020-00341-1
[6]. Perez VM, Kearney JF, Yeh JJ. The PDAC Extracellular Matrix: A Review of the ECM Protein Composition, Tumor Cell Interaction, and Therapeutic Strategies. Front Oncol. 2021;11:751311. doi:10.3389/fonc.2021.751311
[7]. Longo V, Brunetti O, Gnoni A, et al. Angiogenesis in pancreatic ductal adenocarcinoma: A controversial issue. Oncotarget. 2016;7(36):58649-58658. doi:10.18632/oncotarget.10765
[8]. Guo J, Wang S, Gao Q. An integrated overview of the immunosuppression features in the tumor microenvironment of pancreatic cancer. Front Immunol. 2023;14:1258538. doi:10.3389/fimmu.2023.1258538
[9]. Oh K, Yoo YJ, Torre-Healy LA, et al. Coordinated single-cell tumor microenvironment dynamics reinforce pancreatic cancer subtype. Nat Commun. 2023;14(1):5226. doi:10.1038/s41467-023-40895-6
[10]. Fu Y, Liu S, Zeng S, Shen H. The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol Cancer. 2018;17(1):62. doi:10.1186/s12943-018-0815-z
[11]. Jainarayanan A, Mouroug-Anand N, Arbe-Barnes EH, et al. Pseudotime dynamics of T cells in pancreatic ductal adenocarcinoma inform distinct functional states within the regulatory and cytotoxic T cells. iScience. 2023;26(4):106324. doi:10.1016/j.isci.2023.106324
[12]. Goulart MR, Stasinos K, Fincham REA, Delvecchio FR, Kocher HM. T cells in pancreatic cancer stroma. WJG. 2021;27(46):7956-7968. doi:10.3748/wjg.v27.i46.7956
[13]. Saka D, Gökalp M, Piyade B, et al. Mechanisms of T-Cell Exhaustion in Pancreatic Cancer. Cancers. 2020;12(8):2274. doi:10.3390/cancers12082274
[14]. Yang K, Yang T, Yu J, Li F, Zhao X. Integrated transcriptional analysis reveals macrophage heterogeneity and macrophage-tumor cell interactions in the progression of pancreatic ductal adenocarcinoma. BMC Cancer. 2023;23(1):199. doi:10.1186/s12885-023-10675-y
[15]. Poh AR, Ernst M. Tumor-Associated Macrophages in Pancreatic Ductal Adenocarcinoma: Therapeutic Opportunities and Clinical Challenges. Cancers. 2021;13(12):2860. doi:10.3390/cancers13122860
[16]. Zhu Y, Herndon JM, Sojka DK, et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity. 2017;47(2):323-338.e6. doi:10.1016/j.immuni.2017.07.014
[17]. Caronni N, La Terza F, Vittoria FM, et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature. 2023;623(7986):415-422. doi:10.1038/s41586-023-06685-2
[18]. Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer — clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17(9):527-540. doi:10.1038/s41571-020-0363-5
[19]. Zhang J, Song J, Tang S, et al. Multi-omics analysis reveals the chemoresistance mechanism of proliferating tissue-resident macrophages in PDAC via metabolic adaptation. Cell Reports.2023;42(6):112620. doi:10.1016/j.celrep.2023.112620
[20]. Xia Q, Jia J, Hu C, et al. Tumor-associated macrophages promote PD-L1 expression in tumor cells by regulating PKM2 nuclear translocation in pancreatic ductal adenocarcinoma. Oncogene. 2022;41(6):865-877. doi:10.1038/s41388-021-02133-5
[21]. Thomas SK, Wattenberg MM, Choi-Bose S, et al. Kupffer cells prevent pancreatic ductal adenocarcinoma metastasis to the liver in mice. Nat Commun. 2023;14(1):6330. doi:10.1038/s41467-023-41771-z
[22]. Werba G, Weissinger D, Kawaler EA, et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat Commun. 2023;14(1):797. doi:10.1038/s41467-023-36296-4
[23]. Zeng, Pöttler, Lan, Grützmann, Pilarsky, Yang. Chemoresistance in Pancreatic Cancer. IJMS. 2019;20(18):4504. doi:10.3390/ijms20184504
[24]. D’Errico G, Alonso-Nocelo M, Vallespinos M, et al. Tumor-associated macrophage-secreted 14-3-3ζ signals via AXL to promote pancreatic cancer chemoresistance. Oncogene. 2019;38(27):5469-5485. doi:10.1038/s41388-019-0803-9
[25]. Petty AJ, Owen DH, Yang Y, Huang X. Targeting Tumor-Associated Macrophages in Cancer Immunotherapy. Cancers. 2021;13(21):5318. doi:10.3390/cancers13215318
[26]. Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Sig Transduct Target Ther. 2021;6(1):75. doi:10.1038/s41392-021-00484-9
[27]. Fan J qiao, Wang MF, Chen HL, Shang D, Das JK, Song J. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer. 2020;19(1):32. doi:10.1186/s12943-020-01151-3
[28]. De Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41(3):374-403. doi:10.1016/j.ccell.2023.02.016 Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther. 2020;5(1):166. doi:10.1038/s41392-020-00280-x
[29]. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther. 2020;5(1):166. doi:10.1038/s41392-020-00280-x
[30]. Zhang Q, Zhang Z. Stepwise immune alterations in multiple myeloma progression. Nat Cancer. 2020;1(5):477-479. doi:10.1038/s43018-020-0063-1
[31]. Gautam SK, Batra SK, Jain M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol Cancer. 2023;22(1):118. doi:10.1186/s12943-023-01813-y
[32]. Wang R, Song S, Qin J, et al. Evolution of immune and stromal cell states and ecotypes during gastric adenocarcinoma progression. Cancer Cell. 2023;41(8):1407-1426.e9. doi:10.1016/j.ccell.2023.06.005
[33]. Peng J, Sun BF, Chen CY, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725-738. doi:10.1038/s41422-019-0195-y
[34]. Astuti Y, Raymant M, Quaranta V, et al. Efferocytosis reprograms the tumor microenvironment to promote pancreatic cancer liver metastasis. Nat Cancer. Published online February 14, 2024. doi:10.1038/s43018-024-00731-2
[35]. Moncada R, Barkley D, Wagner F, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38(3):333-342. doi:10.1038/s41587-019-0392-8
[36]. Duan Y, Chu H, Brandl K, et al. CRIg on liver macrophages clears pathobionts and protects against alcoholic liver disease. Nat Commun. 2021;12(1):7172. doi:10.1038/s41467-021-27385-3
[37]. Dobie R, Wilson-Kanamori JR, Henderson BEP, et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 2019;29(7):1832-1847.e8. doi:10.1016/j.celrep.2019.10.024
[38]. Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575(7783):512-518. doi:10.1038/s41586-019-1631-3
[39]. Lin W, Noel P, Borazanci EH, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020;12(1):80. doi:10.1186/s13073-020-00776-9
[40]. Steele NG, Carpenter ES, Kemp SB, et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer. 2020;1(11):1097-1112. doi:10.1038/s43018-020-00121-4
[41]. Halbrook CJ, Thurston G, Boyer S, et al. Differential integrated stress response and asparagine production drive symbiosis and therapy resistance of pancreatic adenocarcinoma cells. Nat Cancer. 2022;3(11):1386-1403. doi:10.1038/s43018-022-00463-1
[42]. Lee JJ, Bernard V, Semaan A, et al. Elucidation of Tumor-Stromal Heterogeneity and the Ligand-Receptor Interactome by Single-Cell Transcriptomics in Real-world Pancreatic Cancer Biopsies.
[43]. Clin Cancer Res. 2021;27(21):5912-5921. doi:10.1158/1078-0432.CCR-20-3925 Kemp SB, Steele NG, Carpenter ES, et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci Alliance. 2021;4(6):e202000935. doi:10.26508/lsa.202000935
[44]. Chen K, Wang Q, Liu X, Tian X, Dong A, Yang Y. Immune profiling and prognostic model of pancreatic cancer using quantitative pathology and single-cell RNA sequencing. J Transl Med. 2023;21(1):210. doi:10.1186/s12967-023-04051-4
[45]. Chen J, Liu Z, Wu Z, Li W, Tan X. Identification of a chemoresistance-related prognostic gene signature by comprehensive analysis and experimental validation in pancreatic cancer. Front Oncol. 2023;13:1132424. doi:10.3389/fonc.2023.1132424
[46]. Caronni N, La Terza F, Vittoria FM, et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature. 2023;623(7986):415-422. doi:10.1038/s41586-023-06685-2
[47]. Oh K, Yoo YJ, Torre-Healy LA, et al. Coordinated single-cell tumor microenvironment dynamics reinforce pancreatic cancer subtype. Nat Commun. 2023;14(1):5226. doi:10.1038/s41467-023-40895-6
[48]. Storrs EP, Chati P, Usmani A, et al. High-dimensional deconstruction of pancreatic cancer identifies tumor microenvironmental and developmental stemness features that predict survival. NPJ Precis Oncol. 2023;7(1):105. doi:10.1038/s41698-023-00455-z
[49]. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi:10.1186/s13059-017-1382-0
[50]. Gayoso A, Lopez R, Xing G, et al. A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol. 2022;40(2):163-166. doi:10.1038/s41587-021-01206-w
[51]. Badia-i-Mompel P, Vélez Santiago J, Braunger J, et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Kuijjer ML, ed. Bioinformatics Advances. 2022;2(1):vbac016. doi:10.1093/bioadv/vbac016
[52]. Long Y, Ang KS, Li M, et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat Commun. 2023;14(1):1155. doi:10.1038/s41467-023-36796-3
[53]. Huang Y, Mohanty V, Dede M, et al. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat Commun. 2023;14(1):4883. doi:10.1038/s41467-023-40457-w
[54]. Dimitrov D, Türei D, Garrido-Rodriguez M, et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun. 2022;13(1):3224.
[55]. Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053-1058. doi:10.1038/s41592-018-0229-2
[56]. Mulder K, Patel AA, Kong WT, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54(8):1883-1900.e5. doi:10.1016/j.immuni.2021.07.007
[57]. Cheng S, Li Z, Gao R, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184(3):792-809.e23. doi:10.1016/j.cell.2021.01.010
[58]. Omatsu M, Nakanishi Y, Iwane K, et al. THBS1-producing tumor-infiltrating monocyte-like cells contribute to immunosuppression and metastasis in colorectal cancer. Nat Commun. 2023;14(1):5534. doi:10.1038/s41467-023-41095-y
[59]. Daubon T, Léon C, Clarke K, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun. 2019;10(1):1146. doi:10.1038/s41467-019-08480-y
[60]. Zhang X, Huang T, Li Y, Qiu H. Upregulation of THBS1 is Related to Immunity and Chemotherapy Resistance in Gastric Cancer. IJGM. 2021;Volume 14:4945-4957. doi:10.2147/IJGM.S329208
[61]. Kaur S, Bronson SM, Pal-Nath D, Miller TW, Soto-Pantoja DR, Roberts DD. Functions of Thrombospondin-1 in the Tumor Microenvironment. IJMS. 2021;22(9):4570. doi:10.3390/ijms22094570
[62]. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381-386. doi:10.1038/nbt.2859
[63]. Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13(2):115-124. doi:10.1038/sj.cgt.7700896
[64]. Trizzino M, Zucco A, Deliard S, et al. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci Adv. 2021;7(3):eaaz8836. doi:10.1126/sciadv.aaz8836
[65]. Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The Role of the Transcription Factor EGR1 in Cancer. Front Oncol. 2021;11:642547. doi:10.3389/fonc.2021.642547
[66]. Goodman WA, Omenetti S, Date D, et al. KLF6 contributes to myeloid cell plasticity in the pathogenesis of intestinal inflammation. Mucosal Immunology. 2016;9(5):1250-1262. doi:10.1038/mi.2016.1
[67]. Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289(15):10318-10329. doi:10.1074/jbc.M113.526749
[68]. Kim GD, Das R, Goduni L, McClellan S, Hazlett LD, Mahabeleshwar GH. Kruppel-like Factor 6 Promotes Macrophage-mediated Inflammation by Suppressing B Cell Leukemia/Lymphoma 6 Expression. J Biol Chem. 2016;291(40):21271-21282. doi:10.1074/jbc.M116.738617
[69]. Wang J, Tian Y, Phillips KLE, et al. Tumor necrosis factor α– and interleukin‐1β–dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis & Rheumatism. 2013;65(3):832-842. doi:10.1002/art.37819
[70]. Staversky RJ, Byun DK, Georger MA, et al. The Chemokine CCL3 Regulates Myeloid Differentiation and Hematopoietic Stem Cell Numbers. Sci Rep. 2018;8(1):14691.doi:10.1038/s41598-018-32978-y
[71]. Tsai CH, Lai ACY, Lin YC, et al. Neutrophil extracellular trap production and CCL4L2 expression influence corticosteroid response in asthma. Sci Transl Med. 2023;15(699):eadf3843. doi:10.1126/scitranslmed.adf3843
[72]. Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17(1):38-53. doi:10.1038/nrc.2016.115
[73]. Yi X, Luo L, Zhu Y, et al. SPP1 facilitates cell migration and invasion by targeting COL11A1 in lung adenocarcinoma. Cancer Cell Int. 2022;22(1):324. doi:10.1186/s12935-022-02749-x
[74]. Kuester D, Lippert H, Roessner A, Krueger S. The cathepsin family and their role in colorectal cancer. Pathol Res Pract. 2008;204(7):491-500. doi:10.1016/j.prp.2008.04.010
[75]. Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15(12):712-729. doi:10.1038/nrc4027
[76]. Shi B, Chu J, Huang T, et al. The Scavenger Receptor MARCO Expressed by Tumor-Associated Macrophages Are Highly Associated With Poor Pancreatic Cancer Prognosis. Front Oncol. 2021;11:771488. doi:10.3389/fonc.2021.771488
[77]. Chen Z, Huang Y, Hu Z, et al. Dissecting the single-cell transcriptome network in patients with esophageal squamous cell carcinoma receiving operative paclitaxel plus platinum chemotherapy. Oncogenesis. 2021;10(10):71. doi:10.1038/s41389-021-00359-2
[78]. Sharma A, Seow JJW, Dutertre CA, et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell. 2020;183(2):377-394.e21. doi:10.1016/j.cell.2020.08.040
[79]. Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541-566. doi:10.1146/annurev-physiol-022516-034339
[80]. Xiong A, Zhang J, Chen Y, Zhang Y, Yang F. Integrated single-cell transcriptomic analyses reveal that GPNMB-high macrophages promote PN-MES transition and impede T cell activation in GBM. EBioMedicine. 2022;83:104239. doi:10.1016/j.ebiom.2022.104239
[81]. Yang Q, Lu Y, Shangguan J, Shu X. PSMA1 mediates tumor progression and poor prognosis of gastric carcinoma by deubiquitinating and stabilizing TAZ. Cell Death Dis. 2022;13(11):989. doi:10.1038/s41419-022-05417-0
[82]. Schoeps B, Eckfeld C, Prokopchuk O, et al. TIMP1 Triggers Neutrophil Extracellular Trap Formation in Pancreatic Cancer. Cancer Res. 2021;81(13):3568-3579. doi:10.1158/0008-5472.CAN-20-4125
[83]. Summers HD, Wills JW, Rees P. Spatial statistics is a comprehensive tool for quantifying cell neighbor relationships and biological processes via tissue image analysis. Cell Reports Methods. 2022;2(11):100348. doi:10.1016/j.crmeth.2022.100348
[84]. Qi J, Sun H, Zhang Y, et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat Commun. 2022;13(1):1742. doi:10.1038/s41467-022-29366-6
[85]. Long Y, Ang KS, Li M, et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat Commun. 2023;14(1):1155. doi:10.1038/s41467-023-36796-3
[86]. Huang Y, Mohanty V, Dede M, et al. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat Commun. 2023;14(1):4883. doi:10.1038/s41467-023-40457-w
[87]. Kloosterman DJ, Akkari L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell. 2023;186(8):1627-1651. doi:10.1016/j.cell.2023.02.020
[88]. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889-896. doi:10.1038/ni.1937
[89]. Wang S, Wang J, Chen Z, et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. npj Precis Onc. 2024;8(1):31. doi:10.1038/s41698-024-00522-z
[90]. Zeng W, Li F, Jin S, Ho PC, Liu PS, Xie X. Functional polarization of tumor-associated macrophages dictated by metabolic reprogramming. J Exp Clin Cancer Res. 2023;42(1):245. doi:10.1186/s13046-023-02832-9
[91]. Wang J, Mi S, Ding M, Li X, Yuan S. Metabolism and polarization regulation of macrophages in the tumor microenvironment. Cancer Letters. 2022;543:215766. doi:10.1016/j.canlet.2022.215766
[92]. Sun B, Hyun H, Li L tao, Wang AZ. Harnessing nanomedicine to overcome the immunosuppressive tumor microenvironment. Acta Pharmacol Sin. 2020;41(7):970-985. doi:10.1038/s41401-020-0424-4
[93]. Qiao G, Li S, Pan X, et al. Surgical tumor–derived nanoplatform targets tumor-associated macrophage for personalized postsurgical cancer immunotherapy. Sci Adv. 2024;10(13):eadk7955. doi:10.1126/sciadv.adk7955
[94]. Liu J, Bai Y, Li Y, Li X, Luo K. Reprogramming the immunosuppressive tumor microenvironment through nanomedicine: an immunometabolism perspective. eBioMedicine. 2024;107:105301. doi:10.1016/j.ebiom.2024.105301
Cite this article
Meng,F. (2024). In-depth analysis of pathological function of tumor-associated macrophage populations in hepatic metastatic and primary PDAC. Theoretical and Natural Science,67,84-107.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Halbrook CJ, Lyssiotis CA, Pasca Di Magliano M, Maitra A. Pancreatic cancer: Advances and challenges. Cell. 2023;186(8):1729-1754. doi:10.1016/j.cell.2023.02.014
[2]. Luo W, Wen T, Qu X. Tumor immune microenvironment-based therapies in pancreatic ductal adenocarcinoma: time to update the concept. J Exp Clin Cancer Res. 2024;43(1):8. doi:10.1186/s13046-023-02935-3
[3]. Joseph AM, Al Aiyan A, Al-Ramadi B, Singh SK, Kishore U. Innate and adaptive immune-directed tumour microenvironment in pancreatic ductal adenocarcinoma. Front Immunol. 2024;15:1323198. doi:10.3389/fimmu.2024.1323198
[4]. Zhang S, Fang W, Zhou S, et al. Single cell transcriptomic analyses implicate an immunosuppressive tumor microenvironment in pancreatic cancer liver metastasis. Nat Commun. 2023;14(1):5123. doi:10.1038/s41467-023-40727-7
[5]. Bulle A, Lim KH. Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Sig Transduct Target Ther. 2020;5(1):249. doi:10.1038/s41392-020-00341-1
[6]. Perez VM, Kearney JF, Yeh JJ. The PDAC Extracellular Matrix: A Review of the ECM Protein Composition, Tumor Cell Interaction, and Therapeutic Strategies. Front Oncol. 2021;11:751311. doi:10.3389/fonc.2021.751311
[7]. Longo V, Brunetti O, Gnoni A, et al. Angiogenesis in pancreatic ductal adenocarcinoma: A controversial issue. Oncotarget. 2016;7(36):58649-58658. doi:10.18632/oncotarget.10765
[8]. Guo J, Wang S, Gao Q. An integrated overview of the immunosuppression features in the tumor microenvironment of pancreatic cancer. Front Immunol. 2023;14:1258538. doi:10.3389/fimmu.2023.1258538
[9]. Oh K, Yoo YJ, Torre-Healy LA, et al. Coordinated single-cell tumor microenvironment dynamics reinforce pancreatic cancer subtype. Nat Commun. 2023;14(1):5226. doi:10.1038/s41467-023-40895-6
[10]. Fu Y, Liu S, Zeng S, Shen H. The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol Cancer. 2018;17(1):62. doi:10.1186/s12943-018-0815-z
[11]. Jainarayanan A, Mouroug-Anand N, Arbe-Barnes EH, et al. Pseudotime dynamics of T cells in pancreatic ductal adenocarcinoma inform distinct functional states within the regulatory and cytotoxic T cells. iScience. 2023;26(4):106324. doi:10.1016/j.isci.2023.106324
[12]. Goulart MR, Stasinos K, Fincham REA, Delvecchio FR, Kocher HM. T cells in pancreatic cancer stroma. WJG. 2021;27(46):7956-7968. doi:10.3748/wjg.v27.i46.7956
[13]. Saka D, Gökalp M, Piyade B, et al. Mechanisms of T-Cell Exhaustion in Pancreatic Cancer. Cancers. 2020;12(8):2274. doi:10.3390/cancers12082274
[14]. Yang K, Yang T, Yu J, Li F, Zhao X. Integrated transcriptional analysis reveals macrophage heterogeneity and macrophage-tumor cell interactions in the progression of pancreatic ductal adenocarcinoma. BMC Cancer. 2023;23(1):199. doi:10.1186/s12885-023-10675-y
[15]. Poh AR, Ernst M. Tumor-Associated Macrophages in Pancreatic Ductal Adenocarcinoma: Therapeutic Opportunities and Clinical Challenges. Cancers. 2021;13(12):2860. doi:10.3390/cancers13122860
[16]. Zhu Y, Herndon JM, Sojka DK, et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity. 2017;47(2):323-338.e6. doi:10.1016/j.immuni.2017.07.014
[17]. Caronni N, La Terza F, Vittoria FM, et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature. 2023;623(7986):415-422. doi:10.1038/s41586-023-06685-2
[18]. Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer — clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17(9):527-540. doi:10.1038/s41571-020-0363-5
[19]. Zhang J, Song J, Tang S, et al. Multi-omics analysis reveals the chemoresistance mechanism of proliferating tissue-resident macrophages in PDAC via metabolic adaptation. Cell Reports.2023;42(6):112620. doi:10.1016/j.celrep.2023.112620
[20]. Xia Q, Jia J, Hu C, et al. Tumor-associated macrophages promote PD-L1 expression in tumor cells by regulating PKM2 nuclear translocation in pancreatic ductal adenocarcinoma. Oncogene. 2022;41(6):865-877. doi:10.1038/s41388-021-02133-5
[21]. Thomas SK, Wattenberg MM, Choi-Bose S, et al. Kupffer cells prevent pancreatic ductal adenocarcinoma metastasis to the liver in mice. Nat Commun. 2023;14(1):6330. doi:10.1038/s41467-023-41771-z
[22]. Werba G, Weissinger D, Kawaler EA, et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat Commun. 2023;14(1):797. doi:10.1038/s41467-023-36296-4
[23]. Zeng, Pöttler, Lan, Grützmann, Pilarsky, Yang. Chemoresistance in Pancreatic Cancer. IJMS. 2019;20(18):4504. doi:10.3390/ijms20184504
[24]. D’Errico G, Alonso-Nocelo M, Vallespinos M, et al. Tumor-associated macrophage-secreted 14-3-3ζ signals via AXL to promote pancreatic cancer chemoresistance. Oncogene. 2019;38(27):5469-5485. doi:10.1038/s41388-019-0803-9
[25]. Petty AJ, Owen DH, Yang Y, Huang X. Targeting Tumor-Associated Macrophages in Cancer Immunotherapy. Cancers. 2021;13(21):5318. doi:10.3390/cancers13215318
[26]. Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Sig Transduct Target Ther. 2021;6(1):75. doi:10.1038/s41392-021-00484-9
[27]. Fan J qiao, Wang MF, Chen HL, Shang D, Das JK, Song J. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer. 2020;19(1):32. doi:10.1186/s12943-020-01151-3
[28]. De Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41(3):374-403. doi:10.1016/j.ccell.2023.02.016 Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther. 2020;5(1):166. doi:10.1038/s41392-020-00280-x
[29]. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Sig Transduct Target Ther. 2020;5(1):166. doi:10.1038/s41392-020-00280-x
[30]. Zhang Q, Zhang Z. Stepwise immune alterations in multiple myeloma progression. Nat Cancer. 2020;1(5):477-479. doi:10.1038/s43018-020-0063-1
[31]. Gautam SK, Batra SK, Jain M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol Cancer. 2023;22(1):118. doi:10.1186/s12943-023-01813-y
[32]. Wang R, Song S, Qin J, et al. Evolution of immune and stromal cell states and ecotypes during gastric adenocarcinoma progression. Cancer Cell. 2023;41(8):1407-1426.e9. doi:10.1016/j.ccell.2023.06.005
[33]. Peng J, Sun BF, Chen CY, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725-738. doi:10.1038/s41422-019-0195-y
[34]. Astuti Y, Raymant M, Quaranta V, et al. Efferocytosis reprograms the tumor microenvironment to promote pancreatic cancer liver metastasis. Nat Cancer. Published online February 14, 2024. doi:10.1038/s43018-024-00731-2
[35]. Moncada R, Barkley D, Wagner F, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38(3):333-342. doi:10.1038/s41587-019-0392-8
[36]. Duan Y, Chu H, Brandl K, et al. CRIg on liver macrophages clears pathobionts and protects against alcoholic liver disease. Nat Commun. 2021;12(1):7172. doi:10.1038/s41467-021-27385-3
[37]. Dobie R, Wilson-Kanamori JR, Henderson BEP, et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 2019;29(7):1832-1847.e8. doi:10.1016/j.celrep.2019.10.024
[38]. Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575(7783):512-518. doi:10.1038/s41586-019-1631-3
[39]. Lin W, Noel P, Borazanci EH, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020;12(1):80. doi:10.1186/s13073-020-00776-9
[40]. Steele NG, Carpenter ES, Kemp SB, et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer. 2020;1(11):1097-1112. doi:10.1038/s43018-020-00121-4
[41]. Halbrook CJ, Thurston G, Boyer S, et al. Differential integrated stress response and asparagine production drive symbiosis and therapy resistance of pancreatic adenocarcinoma cells. Nat Cancer. 2022;3(11):1386-1403. doi:10.1038/s43018-022-00463-1
[42]. Lee JJ, Bernard V, Semaan A, et al. Elucidation of Tumor-Stromal Heterogeneity and the Ligand-Receptor Interactome by Single-Cell Transcriptomics in Real-world Pancreatic Cancer Biopsies.
[43]. Clin Cancer Res. 2021;27(21):5912-5921. doi:10.1158/1078-0432.CCR-20-3925 Kemp SB, Steele NG, Carpenter ES, et al. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci Alliance. 2021;4(6):e202000935. doi:10.26508/lsa.202000935
[44]. Chen K, Wang Q, Liu X, Tian X, Dong A, Yang Y. Immune profiling and prognostic model of pancreatic cancer using quantitative pathology and single-cell RNA sequencing. J Transl Med. 2023;21(1):210. doi:10.1186/s12967-023-04051-4
[45]. Chen J, Liu Z, Wu Z, Li W, Tan X. Identification of a chemoresistance-related prognostic gene signature by comprehensive analysis and experimental validation in pancreatic cancer. Front Oncol. 2023;13:1132424. doi:10.3389/fonc.2023.1132424
[46]. Caronni N, La Terza F, Vittoria FM, et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature. 2023;623(7986):415-422. doi:10.1038/s41586-023-06685-2
[47]. Oh K, Yoo YJ, Torre-Healy LA, et al. Coordinated single-cell tumor microenvironment dynamics reinforce pancreatic cancer subtype. Nat Commun. 2023;14(1):5226. doi:10.1038/s41467-023-40895-6
[48]. Storrs EP, Chati P, Usmani A, et al. High-dimensional deconstruction of pancreatic cancer identifies tumor microenvironmental and developmental stemness features that predict survival. NPJ Precis Oncol. 2023;7(1):105. doi:10.1038/s41698-023-00455-z
[49]. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi:10.1186/s13059-017-1382-0
[50]. Gayoso A, Lopez R, Xing G, et al. A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol. 2022;40(2):163-166. doi:10.1038/s41587-021-01206-w
[51]. Badia-i-Mompel P, Vélez Santiago J, Braunger J, et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Kuijjer ML, ed. Bioinformatics Advances. 2022;2(1):vbac016. doi:10.1093/bioadv/vbac016
[52]. Long Y, Ang KS, Li M, et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat Commun. 2023;14(1):1155. doi:10.1038/s41467-023-36796-3
[53]. Huang Y, Mohanty V, Dede M, et al. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat Commun. 2023;14(1):4883. doi:10.1038/s41467-023-40457-w
[54]. Dimitrov D, Türei D, Garrido-Rodriguez M, et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun. 2022;13(1):3224.
[55]. Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15(12):1053-1058. doi:10.1038/s41592-018-0229-2
[56]. Mulder K, Patel AA, Kong WT, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54(8):1883-1900.e5. doi:10.1016/j.immuni.2021.07.007
[57]. Cheng S, Li Z, Gao R, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184(3):792-809.e23. doi:10.1016/j.cell.2021.01.010
[58]. Omatsu M, Nakanishi Y, Iwane K, et al. THBS1-producing tumor-infiltrating monocyte-like cells contribute to immunosuppression and metastasis in colorectal cancer. Nat Commun. 2023;14(1):5534. doi:10.1038/s41467-023-41095-y
[59]. Daubon T, Léon C, Clarke K, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun. 2019;10(1):1146. doi:10.1038/s41467-019-08480-y
[60]. Zhang X, Huang T, Li Y, Qiu H. Upregulation of THBS1 is Related to Immunity and Chemotherapy Resistance in Gastric Cancer. IJGM. 2021;Volume 14:4945-4957. doi:10.2147/IJGM.S329208
[61]. Kaur S, Bronson SM, Pal-Nath D, Miller TW, Soto-Pantoja DR, Roberts DD. Functions of Thrombospondin-1 in the Tumor Microenvironment. IJMS. 2021;22(9):4570. doi:10.3390/ijms22094570
[62]. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381-386. doi:10.1038/nbt.2859
[63]. Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13(2):115-124. doi:10.1038/sj.cgt.7700896
[64]. Trizzino M, Zucco A, Deliard S, et al. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci Adv. 2021;7(3):eaaz8836. doi:10.1126/sciadv.aaz8836
[65]. Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The Role of the Transcription Factor EGR1 in Cancer. Front Oncol. 2021;11:642547. doi:10.3389/fonc.2021.642547
[66]. Goodman WA, Omenetti S, Date D, et al. KLF6 contributes to myeloid cell plasticity in the pathogenesis of intestinal inflammation. Mucosal Immunology. 2016;9(5):1250-1262. doi:10.1038/mi.2016.1
[67]. Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289(15):10318-10329. doi:10.1074/jbc.M113.526749
[68]. Kim GD, Das R, Goduni L, McClellan S, Hazlett LD, Mahabeleshwar GH. Kruppel-like Factor 6 Promotes Macrophage-mediated Inflammation by Suppressing B Cell Leukemia/Lymphoma 6 Expression. J Biol Chem. 2016;291(40):21271-21282. doi:10.1074/jbc.M116.738617
[69]. Wang J, Tian Y, Phillips KLE, et al. Tumor necrosis factor α– and interleukin‐1β–dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis & Rheumatism. 2013;65(3):832-842. doi:10.1002/art.37819
[70]. Staversky RJ, Byun DK, Georger MA, et al. The Chemokine CCL3 Regulates Myeloid Differentiation and Hematopoietic Stem Cell Numbers. Sci Rep. 2018;8(1):14691.doi:10.1038/s41598-018-32978-y
[71]. Tsai CH, Lai ACY, Lin YC, et al. Neutrophil extracellular trap production and CCL4L2 expression influence corticosteroid response in asthma. Sci Transl Med. 2023;15(699):eadf3843. doi:10.1126/scitranslmed.adf3843
[72]. Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17(1):38-53. doi:10.1038/nrc.2016.115
[73]. Yi X, Luo L, Zhu Y, et al. SPP1 facilitates cell migration and invasion by targeting COL11A1 in lung adenocarcinoma. Cancer Cell Int. 2022;22(1):324. doi:10.1186/s12935-022-02749-x
[74]. Kuester D, Lippert H, Roessner A, Krueger S. The cathepsin family and their role in colorectal cancer. Pathol Res Pract. 2008;204(7):491-500. doi:10.1016/j.prp.2008.04.010
[75]. Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer. 2015;15(12):712-729. doi:10.1038/nrc4027
[76]. Shi B, Chu J, Huang T, et al. The Scavenger Receptor MARCO Expressed by Tumor-Associated Macrophages Are Highly Associated With Poor Pancreatic Cancer Prognosis. Front Oncol. 2021;11:771488. doi:10.3389/fonc.2021.771488
[77]. Chen Z, Huang Y, Hu Z, et al. Dissecting the single-cell transcriptome network in patients with esophageal squamous cell carcinoma receiving operative paclitaxel plus platinum chemotherapy. Oncogenesis. 2021;10(10):71. doi:10.1038/s41389-021-00359-2
[78]. Sharma A, Seow JJW, Dutertre CA, et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell. 2020;183(2):377-394.e21. doi:10.1016/j.cell.2020.08.040
[79]. Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541-566. doi:10.1146/annurev-physiol-022516-034339
[80]. Xiong A, Zhang J, Chen Y, Zhang Y, Yang F. Integrated single-cell transcriptomic analyses reveal that GPNMB-high macrophages promote PN-MES transition and impede T cell activation in GBM. EBioMedicine. 2022;83:104239. doi:10.1016/j.ebiom.2022.104239
[81]. Yang Q, Lu Y, Shangguan J, Shu X. PSMA1 mediates tumor progression and poor prognosis of gastric carcinoma by deubiquitinating and stabilizing TAZ. Cell Death Dis. 2022;13(11):989. doi:10.1038/s41419-022-05417-0
[82]. Schoeps B, Eckfeld C, Prokopchuk O, et al. TIMP1 Triggers Neutrophil Extracellular Trap Formation in Pancreatic Cancer. Cancer Res. 2021;81(13):3568-3579. doi:10.1158/0008-5472.CAN-20-4125
[83]. Summers HD, Wills JW, Rees P. Spatial statistics is a comprehensive tool for quantifying cell neighbor relationships and biological processes via tissue image analysis. Cell Reports Methods. 2022;2(11):100348. doi:10.1016/j.crmeth.2022.100348
[84]. Qi J, Sun H, Zhang Y, et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat Commun. 2022;13(1):1742. doi:10.1038/s41467-022-29366-6
[85]. Long Y, Ang KS, Li M, et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat Commun. 2023;14(1):1155. doi:10.1038/s41467-023-36796-3
[86]. Huang Y, Mohanty V, Dede M, et al. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat Commun. 2023;14(1):4883. doi:10.1038/s41467-023-40457-w
[87]. Kloosterman DJ, Akkari L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell. 2023;186(8):1627-1651. doi:10.1016/j.cell.2023.02.020
[88]. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889-896. doi:10.1038/ni.1937
[89]. Wang S, Wang J, Chen Z, et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. npj Precis Onc. 2024;8(1):31. doi:10.1038/s41698-024-00522-z
[90]. Zeng W, Li F, Jin S, Ho PC, Liu PS, Xie X. Functional polarization of tumor-associated macrophages dictated by metabolic reprogramming. J Exp Clin Cancer Res. 2023;42(1):245. doi:10.1186/s13046-023-02832-9
[91]. Wang J, Mi S, Ding M, Li X, Yuan S. Metabolism and polarization regulation of macrophages in the tumor microenvironment. Cancer Letters. 2022;543:215766. doi:10.1016/j.canlet.2022.215766
[92]. Sun B, Hyun H, Li L tao, Wang AZ. Harnessing nanomedicine to overcome the immunosuppressive tumor microenvironment. Acta Pharmacol Sin. 2020;41(7):970-985. doi:10.1038/s41401-020-0424-4
[93]. Qiao G, Li S, Pan X, et al. Surgical tumor–derived nanoplatform targets tumor-associated macrophage for personalized postsurgical cancer immunotherapy. Sci Adv. 2024;10(13):eadk7955. doi:10.1126/sciadv.adk7955
[94]. Liu J, Bai Y, Li Y, Li X, Luo K. Reprogramming the immunosuppressive tumor microenvironment through nanomedicine: an immunometabolism perspective. eBioMedicine. 2024;107:105301. doi:10.1016/j.ebiom.2024.105301









