1.Introduction
Lung cancer is one of the most significant causes of cancer-related death in the world and worldwide cases continue to increase[1]. From 1973 to 2015, the average incidence of lung cancer was 59.0/100,000 person-years, and for survival rate for lung cancer patients is only 10%–20% in 5 years[2]. Lung cancer often disrupts the mucosal immune barriers, such as the bronchial mucosa because of the dysfunction of innate immunity and the lesion of immune cells[3,4]. Among which MAIT cells are a population of innate type T cells and enriched in human mucosal layer[5], indicating its potential role in lung cancer.
Mucosal-associated invariant T (MAIT) cells are a unique subset of innate-like T cells. They play a critical role in immune responses. Unlike conventional T cells that recognize specific antigen peptides, MAIT cells respond to microbial vitamin B metabolites presented by the major histocompatibility complex class I-related molecule, MR1[6]. This allows them to recognize tumors that may attempt to evade immune detection through MHC class I downregulation. MAIT cells are particularly attractive targets for immunotherapeutic strategies due to their abundance in humans:1-10% of T cells in the blood are MAIT cells, and they constitute up to 45% of liver lymphocytes[7].
Activated MAIT cells can produce a wide array of cytokines, such as tumor necrosis factor-alpha (TNFa) , perforin and IL-17, which can influence cancer development[8,9]. Moreover, MAIT cells exhibit indirect anti-cancer activity through Lymphokine-Activated Killer (LAK) activity mediated by IFN-y[10] and can release perforin after recognizing MR1+ target cells, thereby mediating pathogen clearance. The potential of MAIT cells becomeing new killer cells in mucosal oncoimmunity makes them a focus of investigation to determine their role in lung cancer inhibition.
Recent studies have shown the significant role of unconventional T cells, such as yδ T cells, natural killer (NK) T cells, and MAIT cells, in regulating tumor progression[11,12]. For instance, MAIT cells have been shown to regulate the progression of colorectal cancer (CRC) by producing INF-y, which can induce tumor cell cycle arrest and apoptosis via the p53 signaling pathway[11]. They can also activate other immune cells, such as NK cells, T cells, and macrophages, thus indirectly contributing to antitumor immunity. In hepatocellular carcinoma (HCC), MAIT cells have been shown to play a critical role in anti-tumor immunity, with studies in wild-type (WT) mice showing MAIT-deficient Mr1−/− mice presented with higher tumor burden[13].
Interestingly, whether MAIT cells interacting with MR1 on tumor cells promote cancer or contribute to anticancer immunity is still debated. For instance, in colorectal cancer patients, increased tumor infiltration of MAIT cells correlates with poor survival due to the production of IL-17, which promotes tumor growth and metastasis[14]. Another research shows similar result on mouse melanoma cell lines B16F10 , which MAIT cells can interact with MR1 on tumor cells to suppress NK cell function, therefore promoting cancer metastasis[15]. This suggests that blocking MR1 or saturating it with an inhibitory ligand could represent a new therapeutic strategy for cancer.
Despite these findings, the immunological mechanisms, distribution, and function of MAIT cells in lung cancer remain unclear. And because of the specificity of MR1 in antigen presentation and its relatively low side effects, approaches that target MR1 and MAIT cells in disease treatment become especially attractive. This study aims to explore the possible relationship between MAIT cells and lung cancer, focusing on their interaction with MR1 molecules on tumor cells by investigating how the presence or absence of MR1 influences MAIT cell proliferation and cytokine production, including IFN-y, perforin, and IL-10, in lung cancer. The goal is to understand the potential relationship between MAIT cells and lung cancer and explore new therapeutic avenues involving MR1 and MAIT cells.
2.Methods
2.1.MRI Overexpression Line
2.1.1.Cell Passage and Activation. Lung metastatic carcinoma (A9) cells (which have low MR1 expression), TC1 (TC-1 murine lung epithelial carcinoma, which highly expresses MR1) cells, and MAIT T cells were obtained and activated by incubating them at 37°C for three days. The cells were then cultivated in DMEM culture medium at a density of 5 times 1×106 cells/mL, with 10%-20% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/mL streptomycin. The air contained 5% of CO2. After three days, the cells were passaged respectively.
2.1.2.Clone MR1 gene. To amplify the MR1 gene from TC1 cells:
First, total RNA was extracted using Trizol. Next, reverse transcription was performed with the HiScript® II 1st Strand cDNA.
Synthesis Kit (cat# K1621, Vazyme), following these steps: add 8 µL ddH₂O, 2 µg total RNA, heat at 65°C for 5 minutes. Secondly, add 2 µL gDNA wiper mix, incubate at 42°C for 2 minutes. Then add 2 µL RT mix, 2 µL HiScript II Enzyme Mix, 1 µL Oligo dt20 VN, 5 µL H₂O, and use a thermal cycler (25°C for 5 minutes, 37°C for 45 minutes, 85°C for 6 seconds) for 35 cycles, to synthesize cDNA. Next, PCR was performed using Champagne Taq DNA Polymerase(cat # P122-d1/d2, Vazyme)to select and amplify the MR1 gene with the following primers:
- Forward: 5'-gattctagagctagctacccccttgactaccgcaag-3'
- Reverse: 5'-atccttcgcggccgctcatcgatctggtgttggaag-3'
To verify the PCR result, the Gel Electrophoresis has been down. 0.5 g agarose was dissolved in 50 mL 1×TAE buffer by microwaving until boiling. After cooling to 60°C, SafeGreen dye(cat# S33112, Vazyme) was added.
Samples were prepared by mixing 100 ng DNA(5ul) with 2 µL loading buffer. DNA marker DL10000 was loaded into the first well, and the gel was run at 120V. The gel was observed under UV light.
2.1.3.Plasmid Construction. After obtaining the MR1 PCR product, both the MR1 gene and the pCDH-GFP-Puro plasmid were digested with EcoRI and BamHI. Reaction mixtures for digestion included: Plasmid DNA: 2 µL plasmid DNA, 15 µL water, 2 µL 10× FastDigest buffer(cat# ER1461, Thermoscientific), 1 µL FastDigest enzyme. and PCR product: 10 µL PCR product, 17 µL water, 2 µL 10x FastDigest buffer, 1 µL FastDigest enzyme.
Then, the digestion products were combined: 1 µL from each digestion reaction, 4 µL 5x CE MultiS Buffer, 2 µL Exnase MultiS(cat# c112-01, Vazyme),and 10 µL ddH₂O, incubated at 37°C for 30 minutes, then placed on ice. The recombinant plasmid was introduced into competent E. coli cells(cat # C4040-03, Thermofisher), which were incubated on ice for 30 minutes, heat-shocked at 42°C for 45 seconds, and incubated on ice for 3 minutes. Next,900 µL LB medium was added, and the cells were incubated at 37°C for 1 hour, then centrifuged, and plated on LB agar for overnight incubation at 37°C. Plasmids were extracted using PureLink Quick Plasmid Miniprep Kit(cat # K210010, Invitrogen).
2.1.4.Transfection. The pCDH-GFP-Puro-MR1 plasmid was transfected into A9 cells using Lipo6000(cat# C0526, Invitrogen,). 30 µL Lipo6000 and 750 µL Opti-MEM were mixed in one tube, and 15 µg plasmid and 750 µL Opti-MEM were mixed in another. The mixtures were combined and incubated for 48 hours. Use Flow Cytometry analysis to sout EGFP-positive cells to obtain successful transfectant cell .
2.2.MR1 lowexpression line
2.2.1.CRISPR-Cas9 knockout MR1. To knockdown MR1 in TC1 cells, the pspCas9(BB)-2A-Puro (PX459) plasmid was cut with BbsI. sgRNA targeting MR1 in TC1 cells was annealed and ligated into the plasmid. Recombinant plasmid was introduced into competent E. coli, which were incubated on ice for 30 minutes, heat-shocked at 42°C for 45 seconds, and incubated on ice for 3 minutes. Next,900 µL LB medium was added, and the cells were incubated at 37°C for 1 hour, then centrifuged, and plated on LB agar for overnight incubation at 37°C. Plasmids were extracted with PureLink Quick Plasmid Miniprep Kit (Invitrogen, cat # K210010).and transfected into TC1 cells using Lipo6000(cat#C0526, Invitrogen). Use the FLag tag monoclones (10 monoclones) to select the successful MR1 knockout cell.
2.2.2.Western Blot. To verify the success of transfection and knockdown, Cells were lysed using RIPA Lysis Buffer System (sc-24948, Santa Cruz Biotechnology) with PMSF, centrifuged at 12,000 rpm for 10-15 minutes at 4°C, and the supernatant was collected.
Protein concentration was determined using the BCA assay. Samples were denatured at 95°C for 5 minutes, loaded onto a gel, and electrophoresed at 80-120V to let proteins transfer to a membrane. Then blocked with 5% skim milk, and incubated with rabbit anti-mouse MR1(Thermofisher, cat # 13260-1-AP; Abcam, cat # ab229987) polyclonal antibodies. The membrane was washed with TBST, incubated with secondary antibody, and visualized with ECL(LAS-4000).
2.3.MAIT proliferate and cytokines product detection
2.3.1.Flow cytometry analysis. To detect MAIT T cell proliferation after incubate with different groups of cell,Firstly,MAIT sample were Remove supernatant and add CellTraceTM CFSE 1:1000 dilution (cat #C34554, C34570 ),and incubated at 37°C for 20 minutes,follow by adding cold (4°C) complete medium to stop the CFSE reaction.Perform three washes with cold PBS to remove excess CFSE. Then add complete culture medium and mix,Incubate at 37°C for 5 minutes.
After that, seven samples were incubated with dyed MAIT for totally 11 days, they are: control group only with MAIT cell(IK-C1 cell line), A9 with MAIT, high express MR1 A9 with MAIT, high express MR1 A9 with MR1 antibody with MAIT ,only TC1 with MAIT,TC1 knockout with MAIT, TC1 with MR1 antibody with MAIT. Human rIL-2 (5 IU/ml; Peprotech) was added at day 5 and thereafter every 2 d.
After that at day 11, resuspend the cell pellet in fresh, pre-warmed complete culture medium. Pellet the cells and remove the supernatant. Finally analyzed by flow cytometry.(CytoFLEX s beckmen coulter).
2.3.2.ELISA workflow. To measure cytokine and protein production (IL-10, perforin, INF-y) after co-incubation of cells with MAIT T cells: Seven Samples were prepared and incubated. There are 7 groups: control group only with MAIT cell(IK-C1 cell line), A9 with MAIT, high express MR1 A9 with MAIT, high express MR1 A9 with MR1 antibody with MAIT ,only TC1 with MAIT,TC1 knockout with MAIT, TC1 with MR1 antibody with MAIT. Detection antibody and Streptavidin-HRP were added, followed by TMB solution and Stop Solution(Thermofishercat# 88-7316).Absorbance was measured at 450 nm using PerkinElmer EnVision 2030 Multilabel Plate Reader.
2.4.Statistical analysis
Statistical Analysis: Data were analyzed using GraphPad Prism software. Statistical significance was determined using ANOVA or t-tests where appropriate, with p-values < 0.05 considered significant.
3.Results
3.1.The evaluation of transfection efficiency
Following the transfection of MR1 into A9 cells using Lipo6000, the efficiency of transfection was evaluated by observing the EGFP-positive A9 cell under inverted fluorescence microscope to identify the optimal transfection clone. Two wells containing identically transfected A9 cells were analyzed with four different plasmid concentrations(1ul/15ug, 2ul/25ug, 4ul/50ug, 8ul/100ug). As shown in Figure 1, both wells show no EGFP expression under concentration of 1 ul/15ug, as no green fluorescence was visualized. At a concentration of 2 uL/25 ug, some green fluorescence was observed, suggesting that a small amount of plasmid was successfully transfected into the cells. The highest transfection efficiency was observed at a concentration of 4 uL/50 ug, indicated by a significant number of green fluorescent cells. However, at a concentration of 8 uL/100 ug, the plasmid was toxic to the cells, resulting in no viable data. Based on the observation, the 4 ul/50ug concentration cell was selected for subsequent analyses, including gel and Western detection to further confirm successful transfection.
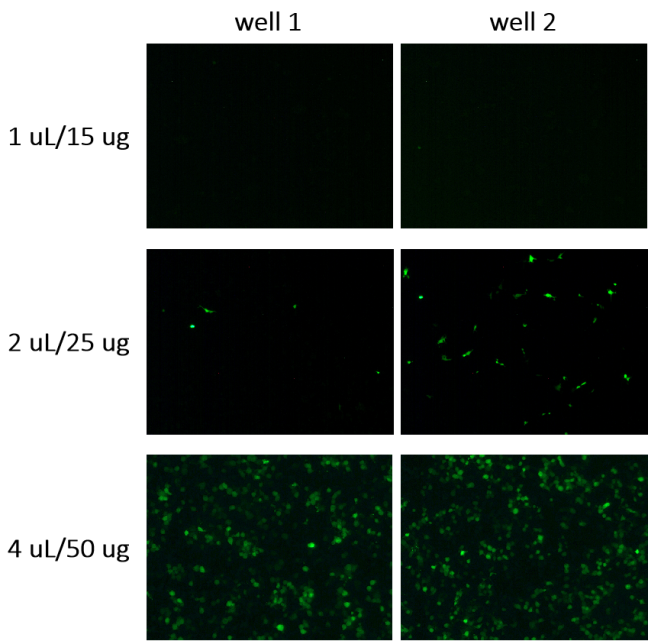
Figure 1. Observation under the inverted fluorescence microscope of EGFP-positive A9 cell. After transfecting pCDH-GFP-Puro-MR1 plasmid using Lipo6000, cells were incubated at 37°C for 48 hours. Transfection efficiency was evaluated according to GFP fluorescence intensity. No data were shown for 8ul/100 ug as the toxicity.
3.2.Gel electrophoresis and Western blot detection
To assess the effectiveness of Cas-9-mediated MR1 knockout in TC1 cells, gel electrophoresis was performed. 10 monoclones are shown in Figure 2. The MR1 in groups 1, 6, 8, and 10 were unsuccessfully knockout and exhibited strong MR1 expression as the band showed prominence at 1025 bp. Groups 2 and 4 showed weaker bands, indicating partial knockout of MR1. Groups 3 and 7 had faint bands, suggesting a near-complete knockout with minimal MR1 remaining. In contrast, groups 5 and 9 showed no detectable MR1 band, indicating a complete and successful knockout. These two groups(5 and 9) were selected for further analysis, including ELISA and Flow Cytometry analysis.
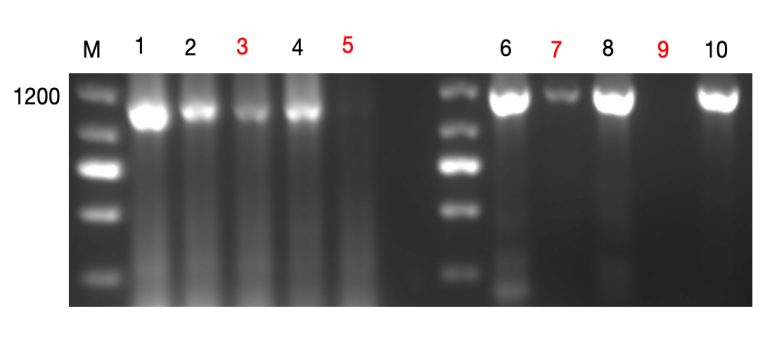
Figure 2. Gel electrophoresis for detecting MR1 knockout in TC1 cell using cas-9 plasmid.The 100 ng DNA (5ul) and 2 µL loading buffer were mixed, and the gel was run at 120V. The results were observed under UV light. The molecular weight of MR1 is 1025bp. Lanes 1-10 represent individual monoclones lines (10 monoclones), with lanes 3 and 7 TC1 cell showing nearly-complete knockout MR1 and 5 and 9 complete knockout. Lane M represents DNA marker.
In Figure 3, Western blot was used to verify MR1 transfection and knockout in five samples(A9 overexpressing MR1, A9, TC1 , TC1 knocking-outMR1 and β-actin is used as the control).The β-actin bands confirmed consistent protein loading across all samples. MR1 protein expression was observed in both A9 cells overexpressing MR1 and in TC1 cells, indicating successful transfection in these groups. In contrast, A9 and MR1-knockout TC1 cells did not express MR1, confirming successful knockout in these groups. These results validate that the samples were prepared correctly and are suitable for subsequent ELISA and Flow Cytometry analysis assays.
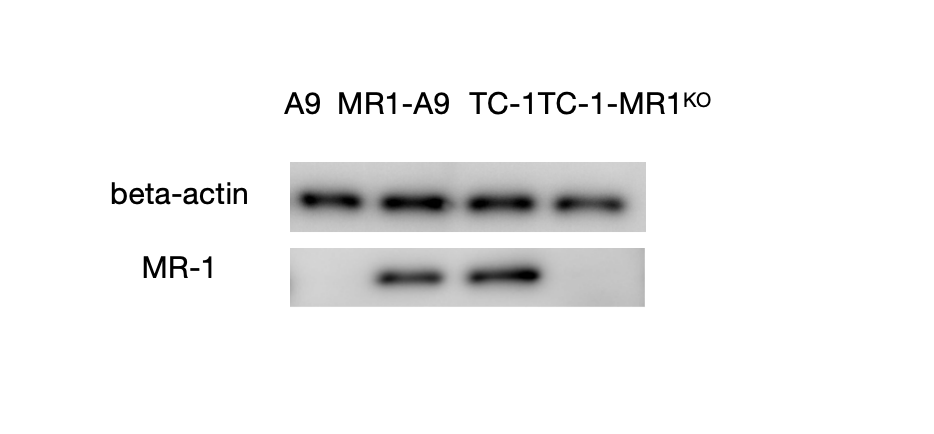
Figure 3. Western Blot Analysis of MR1 Expression in five Cell Groups: Western blot was used to assess the MR1 expression levels of five groups: beta-actin serves as the control, A9 cell line, A9 overexpresses MR1, TC1 , and TC1 knockouts MR1. Using the RIPA Lysis Buffer System, four cell groups were lysed, and the extracted proteins were separated by SDS-PAGE and transferred onto a membrane. The membrane was then probed with an MR1-specific antibody, and protein bands were visualized using X-ray film after a 24-hour incubation period. The membrane was then probed with an MR1-specific antibody, and protein bands were visualized using X-ray film after a 24-hour incubation period.
3.3.Flow Cytometry analysis of proliferation of MAIT
Flow Cytometry analysis was conducted to evaluate the proliferation of MAIT cells across seven different experimental groups, with a focus on understanding the role of MR1 in T cell activation.
In the control group, which consisted solely of MAIT cells, a minimal proliferation rate of 0.97% was observed. The number of proliferation times is estimated to be 1 or less as indicated by a single peak on the Flow Cytometry analysis histogram. This low proliferation suggests limited activation in the absence of additional stimuli.
The group MR1-A9 and group TC-1,which both had high expression of MR1, had the proliferation rate of MAIT 28.1% and 41.4% respectively. In TC-1 group, MAIT proliferated 2 times and in MR1-A9 group, it poliferated 1 times. This robust proliferation underscores the critical role of MR1 in enhancing T cell activation, as both groups involving MR1 showed the highest levels of cell division.
In contrast, the groups without MR1 (A9 group and TC-1-MR1KO group) showed different result. MAIT only proliferate 9.41% in A9 group and 9.05% in TC-1-MR1KO group. None of those groups showed significant proliferation peaks, so MAIT in both group proliferated only 1 time or less.
Finally, in group MR1-A9+Ab group and TC-1+Ab group, MAIT showed low proliferation rate, which were 8.56% and 19.6% respectively, and MAIT proliferated less than one time.
In conclusion, the data indicate that MR1 plays a pivotal role in promoting MAIT cell proliferation, with the highest rates observed in the A9 with MR1 and T-C1 groups.
Figure 4. Flow Cytometry analysis of tumer MR1 toMAIT proliferation. Use Flow Cytometry analysis to detect The influence of tumor MR1 to T cell activation. There are 7 groups co-cultured with MAIT: control group with only IK-C1 cell line, A9 , high express MR1 on A9 (A9-MR1), high express MR1 A9 with MR1 antibody (A9-MR1+ab), TC-1 ,TC-1-MR1KO ,TC1 with MR1 antibody(TC-1+Ab group)
a) The bars represent the cell proliferation. X axis is light intensity and y is number of cells, illustrating the extent of MAIT cell proliferation under different conditions.
Samples were stained with CellTrace CFSE (Carboxyfluorescein Succinimidyl Ester) incubated at 37°C for 20 minutes, and analyzed by flow cytometry. The data of percentage was analysed by Flowjo
b)Bar chart at left It shows the persentage of MAIT that proliferate. The table at right showed all the persentages of three replicates experiments for each group.
3.4.Detection of cytokines and protein by ELISA
To assess the T cell response to MR1, two types of cytokines and a protein produced by T cells in seven different groups were analyzed using ELISA.
IFN-γ ,a crucial cytokine in anti-tumor immunity, could arrest cell cycle on cancer cell by regulating the p53 signaling[11] and acts as a cytotoxic cytokine together with granzyme B and perforin to initiate apoptosis in tumor cells[16]. The control group, which contained only T cells, produced the lowest concentration of IFN-γ (90 pg/ml). The A9 , A9-MR1+ab, TC-1-MR1KO, and TC-1+Ab group all produced similar concentrations, around 150 pg/ml. The highest concentration of IFN-γ was observed in the TC1 group (390 pg/ml), followed closely by A9-MR1 group (380 pg/ml).
IL-10 was initially defined as “cytokine synthesis inhibitory factor”, and in cancer, IL-10 stimulate tumor growth by activating STAT3.The second graph in Figure 4a (IL-10) showed a similar trend. T cells in the TC1 group produced the most IL-10 (440 pg/ml), with the A9-MR1 group being the second highest (280 pg/ml). The other five groups had lower concentrations, fluctuating around 200 pg/ml.
Perforin is an essential molecule in cytotoxic T cell-mediated lysis of target cells . In the third graph of Figure 4a (Perforin), T cells in the TC1 group again produced the highest concentration (780 pg/ml), followed by the A9-MR1 group (520 pg/ml). The other five groups had significantly lower concentrations, around 380 pg/ml.
Overall, the A9-MR1 and TC1 groups exhibited the most robust T cell responses, as indicated by the elevated levels of IFN-γ, IL-10, and perforin.
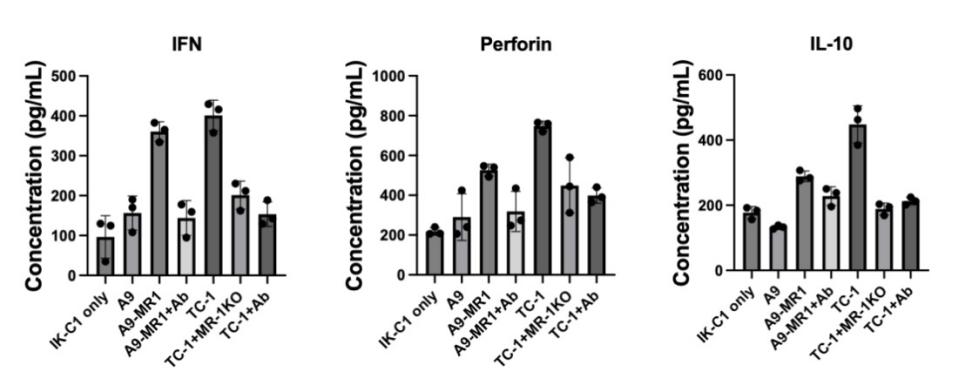
Figure 5. Detection of the influence of tumor MR1 to T cell activation using ELISA. groups are: control group only with MAIT cell(IK-C1 cell line), A9 with MAIT, high express MR1 A9 with MAIT, high express MR1 A9 with MR1 antibody with MAIT ,only TC1 with MAIT,TC1 knockout with MAIT, TC1 with MR1 antibody with MAIT. Incubated samples with capture antibody(for three cytokines), blocked, and washed, add Detection antibody(biotin conjugate), finally add Stop Solution, Absorbance was measured at 450 nm. Repeat doing 3 times. Y axis is cytokine’s concentration.
4.Discussion & Conclusion
Lung cancer (LC) is one of the leading causes of tumor mortality around the world with a persistently low patients survival rate[17]. Understanding the immune mechanisms underlying lung cancer is crucial for developing new therapeutic strategies. The tumor microenvironment of lung cancer is notably complex, incorporating various immune evasion mechanisms[18].
Some recent research shows that anti-PD-1 therapy or immune checkpoint blockade (ICB) with antibodies to PD-1 or PD-L1 could reinvigorate MAIT cells’ antitumor effector functions and therefore improved overall Survival in melanoma patients and non-small cell lung cancer (NSCLC)[19,20]. From the research, the reseptor on the surface of cell plays an important role on how MAIT cell response to cancer.
This study aims to explore the interactions between mucosal-associated invariant T (MAIT) cells and lung cancer cells, particularly focusing on how MR1 expression influences MAIT cell function. Our findings reveal that MAIT cells have a dual role in tumor immunity, which could provide novel insights into lung cancer treatment strategies.
In this study, we evaluated the interaction of MAIT cells with MR1-positive lung cancer cell (TC-1 and MR1-A9) or MR1-negative cancer cell (TC-1-MR1KO and A9) in vitro, focusing on MAIT division, IFN-γ, Perforin, and IL-10 production. Our results provide new insights of the dual role of MAIT cells in tumor immunity and provide a basis for potential therapeutic interventions.
Our results demonstrate that MAIT cells proliferate significantly upon interaction with MR1-positive lung cancer cells (as seen in the TC-1 and MR1-A9), but not with MR1-negative cells (TC-1-MR1KO) (Fig4). This suggests that MR1 plays a role in the activation and proliferation of MAIT cells , reinforcing its function as an antigen-presenting molecule[6]. The MR1-dependent activation of MAIT cells suggests a potential pathway for enhancing immune surveillance in lung cancer.
IFN-γ is known to arrest the cell cycle in cancer cells. One of the function is regulating the p53 signaling pathway and to induce apoptosis through its cytotoxic effects alongside granzyme B and perforin[11,16]. Studies have proven that IFN-γ has anti- cancer function in different type of cancer[11,12,21]. In Lewis lung carcinoma, IFN-γ is shown to crosstalk with M1-like immunostimulatory tumor-associated macrophages (TAMs) to promote anti-tumor activity[12]. In our study, MAIT cells showed a significantly increase in IFN-γ secretion when co-cultured with MR1-positive lung cancer cells (Figure 5). This further confirm the potential anti-tumor function of MAIT cells in immune surveillance of lung cancer. Our finding is consistant with the research on CRC patients,as they also find that activated MAIT cells produce high level of INF[11].
Perforin creates pores in the membrane of target cells, allowing granzyme B to enter and induce apoptosis[22]. Although the role of MR1 in promoting perforin production in lung cancer has been understudied, our experimental results show that MAIT cells release high levels of perforin when contacting MR1-positive lung cancer cells(group MR1-A9 and group TC-1), showing its cytotoxicity. This novel observation indicates that MAIT cells may directly kill lung cancer cells, especially in the case of MR1 antigen presentation, offering a new avenue for research into their cytotoxic functions in tumor microenvironment.
Despite their potential anti-tumor activity, our study also found that MAIT cells produce higher levels of IL-10 when interacting with MR1-positive lung cancer cells. While IL-10 has immunoregulatory functions and can suppress inflammation[23,24,25],its overexpression in the tumor microenvironment may lead to immune escape and tumor progression by activating STAT3 and inhibiting the dendertic cell function[26]. This finding suggests that MAIT cells may have a complex dual role in tumor immunity, which can either be anti-tumor or promote tumor growth through the secretion of IL-10. Other studies, such as those on melanoma cell lines B16F10,as mentioned before, have shown that MAIT cells can interact with MR1 on tumor cells to suppress NK cell function, therefore promoting cancer metastasis[15] further prove our thought. Notably, very few studies have tested the IL-10 production by MAIT associated with MR1 in lung cancer, so our research gives a new sight of the function of MAIT that might promote lung cancer, and this might be a key area for future investigation.
Our results are consistent with findings in other cancers research, such as hepatocellular carcinoma and colorectal cancer, where MAIT cells play different role in cancer, even in same cancer type. For instance, MAIT shows immunosuppressive function in some types of cancer. As mentioned before, MAIT produce IL-17 ,which is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma[14,27,28]. And similarly in HCC, MAIT cells are present with an activating exhausted phenotype, related to tumor promotion[29,30].However, in some cases, it shows antitumor activity instead, for example it produce high level of IFN-y in colorectal cancer[11],which may activate Macrophage and enhance DC maturation. MAIT also mediated pronounced and consistent antitumor activity and prolonged survival in liver tumors, lung metastases, and subcutaneous tumors[31].
This duality may depend on specific conditions within the tumor microenvironment, such as the expression level of MR1, cytokines secreted by tumor cells, different immune context or stages of cancer progression. Further research into how these factors influence MAIT cell behavior in lung cancer is essential.
While our study provides valuable insights into the role of MAIT cells in lung cancer, there are several limitations to consider. The intricacy of the tumor microenvironment in vivo cannot be completely replicated by in vitro models, like the ones used in this study. To gain a deeper understanding of the interactions between immune components and MAIT cells inside the lung cancer microenvironment, animal models should be used in future study. Additionally, expanding the range of cytokines analyzed, such as IL-17, could provide a more comprehensive understanding of the dual functions of MAIT cells. Exploring ways to enhance the anti-tumor activity of MAIT cells by regulating MR1 expression or blocking IL-10 could offer new therapeutic avenues.
In conclusion, This study highlights the complex role of MAIT cells in lung cancer, particularly their dual role in MR1-positive tumors. Our findings clarify the relationship between MR1 and MAIT cells, opening up new avenues for immunotherapy research, especially in terms of methods that might boost anti-tumor responses by controlling MAIT cell activity. regarding. This complexity suggests that the tumor microenvironment plays a significant role in determining the functional outcomes of MAIT cell activity. Therefore, understanding how to manipulate these factors could be key to developing effective immunotherapies targeting MAIT cells in lung cancer. Further research into the regulation of MAIT cell functions could lead to innovative therapies that improve outcomes for lung cancer patients.
References
[1]. Oliver AL. Lung cancer: Epidemiology and screening. Surg Clin North Am (2022) 102(3):335–44. 10.1016/j.suc.2021.12.001
[2]. Lu T, Yang X, Huang Y, Zhao M, Li M, Ma K, et al.. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag Res (2019) 11:943–53. 10.2147/CMAR.S187317
[3]. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20(1):105.
[4]. Quaratino S, Forssmann U, Marschner JP. New approaches in immunotherapy for the treatment of lung cancer. Curr Top Microbiol Immunol (2017) 405:1–31. doi: 10.1007/82_2014_428
[5]. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20(9):1110
[6]. Reantragoon R. et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. The Journal of experimental medicine. 209, 761–774 (2012).
[7]. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al.. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. 10.1182/blood-2010-08-303339
[8]. Yang B, Kang H, Fung A, Zhao H, Wang T, Ma D. The role of interleukin 17 in tumour proliferation, angiogenesis, and metastasis. Mediators Inflamm. (2014) 2014:623759. 10.1155/2014/623759
[9]. Raval A, Puri N, Rath PC, Saxena RK. Cytokine regulation of expression of class I MHC antigens. Exp Mol Med.(1998) 30:1–13. 10.1038/emm.1998.1
[10]. Eun Jeong Won, #1 Jae Kyun Ju, #2 Young-Nan Cho, #3 Hye-Mi Jin, 3 Ki-Jeong Park, 3 Tae-Jong Kim, 3 Yong-Soo Kwon, 4 Hae Jin Kee, 5 Jung-Chul Kim, 2 Seung-Jung Kee, 1 and Yong-Wook Park3Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer2016 Nov 15; 7(46): 76274–76290.
[11]. Ling L, Lin Y, Zheng W, Hong S, Tang X, Zhao P, et al.. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci Rep. (2016) 6:20358. 10.1038/srep20358
[12]. Jorgovanovic, D., Song, M., Wang, L., & Zhang, Y. (2020). Roles of IFN-γ in tumor progression and regression: A review. Biomarker Research, 8(1). https://doi.org/10.1186/s40364-020-00228-x
[13]. Ruf, B., Bruhns, M., Babaei, S., Kedei, N., Ma, L., Revsine, M., Benmebarek, M., Ma, C., Heinrich, B., Subramanyam, V., Qi, J., Wabitsch, S., Green, B. L., Bauer, K. C., Myojin, Y., Greten, L. T., McCallen, J. D., Huang, P., Trehan, R., … Greten, T. F. (2023). Tumor-associated macrophages trigger MAIT cell dysfunction at the HCC invasive margin. Cell, 186(17), 3686-3705.e32. https://doi.org/10.1016/j.cell.2023.07.026
[14]. Zabijak L, Attencourt C, Guignant C, Chatelain D, Marcelo P, Marolleau J-P, et al.. Increased tumor infiltration by mucosal-associated invariant T cells correlates with poor survival in colorectal cancer patients. Cancer Immunol Immunother. (2015) 64:1601–8. 10.1007/s00262-015-1764-7
[15]. Yan, J, Allen, S, McDonald, E, Das, I, Mak, JYW, Liu, L, Fairlie, DP, Meehan, BS, Chen, Z, Corbett, AJ, Varelias, A, Smyth, MJ, Teng, MWL. 2020. MAIT Cells Promote Tumor Initiation, Growth, and Metastases via Tumor MR1. Cancer Discovery. 10:124-141. doi: 10.1158/2159-8290.CD-19-0569. https://www.ncbi.nlm.nih.gov/pubmed/31826876.
[16]. Maimela NR, Liu S, Zhang Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput Struct Biotechnol J. 2018;(xxxx):1–13.
[17]. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
[18]. Zhou, Z., Wang, J., Wang, J., Yang, S., Wang, R., Zhang, G., Li, Z., Shi, R., Wang, Z., & Lu, Q. (2024). Deciphering the tumor immune microenvironment from a multidimensional omics perspective: Insight into next-generation CAR-T cell immunotherapy and beyond. Molecular Cancer, 23(1). https://doi.org/10.1186/s12943-024-02047-2
[19]. Vorwald, V. M., Davis, D. M., Van Gulick, R. J., Torphy, R. J., Borgers, J. S., Klarquist, J., Couts, K. L., Amato, C. M., Cogswell, D. T., Fujita, M., Castleman, M. J., Davis, T., Lozupone, C., Medina, T. M., Robinson, W. A., Gapin, L., McCarter, M. D., & Tobin, R. P. (2022). Circulating CD8<sup>+</sup> mucosal‐associated invariant T cells correlate with improved treatment responses and overall survival in anti‐PD‐1‐treated melanoma patients. Clinical & Translational Immunology, 11(1). https://doi.org/10.1002/cti2.1367
[20]. Sundström, P., Dutta, N., Rodin, W., Hallqvist, A., Raghavan, S., & Quiding Järbrink, M. (2024). Immune checkpoint blockade improves the activation and function of circulating mucosal-associated invariant T (MAIT) cells in patients with non-small cell lung cancer. OncoImmunology, 13(1). https://doi.org/10.1080/2162402x.2024.2312631
[21]. Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16(3):131–144. doi: 10.1038/nrc.2016.14.
[22]. Lopez, J. A., Susanto, O., Jenkins, M. R., Lukoyanova, N., Sutton, V. R., Law, R. H. P., ... & Browne, K. A. (2013). "Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack." Blood, 121(14), 2659-2668.
[23]. Wilke, C. M., Wei, S., Wang, L., Kryczek, I., Kao, J., & Zou, W. (2011). Dual biological effects of the cytokines interleukin-10 and interferon-γ. Cancer Immunology, Immunotherapy, 60(11), 1529-1541. https://doi.org/10.1007/s00262-011-1104-5
[24]. Mocellin S, Panelli M, Wang E, Rossi CR, Pilati P, Nitti D, et al.. IL-10 stimulatory effects on human NK cells explored by gene profile analysis. Genes Immunity. (2004) 5:621–30. 10.1038/sj.gene.6364135
[25]. Park JY, Lee SH, Yoon SR, Park YJ, Jung H, Kim TD, et al.. IL-15-induced IL-10 increases the cytolytic activity of human natural killer cells. Mol Cells. (2011) 32:265–72. 10.1007/s10059-
[26]. Carlini, V., Noonan, D. M., Abdalalem, E., Goletti, D., Sansone, C., Calabrone, L., & Albini, A. (2023). The multifaceted nature of IL-10: Regulation, role in immunological homeostasis and its relevance to cancer, COVID-19 and post-COVID conditions. Frontiers in Immunology, 14. https://doi.org/10.3389/fimmu.2023.1161067
[27]. Shaler, C. R., Tun-Abraham, M. E., Skaro, A. I., Khazaie, K., Corbett, A. J., Mele, T., Hernandez-Alejandro, R., & Haeryfar, S. M. (2017). Mucosa-associated invariant T cells infiltrate hepatic metastases in patients with colorectal carcinoma but are rendered dysfunctional within and adjacent to tumor microenvironment. Cancer Immunology, Immunotherapy, 66(12), 1563-1575. https://doi.org/10.1007/s00262-017-2050-7
[28]. Liu J, Duan Y, Cheng X, et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochemical and Biophysical Research Communications. 2011;407(2):348–354.
[29]. Berzins, S. P., Wallace, M. E., Kannourakis, G., & Kelly, J. (2020). A role for MAIT cells in colorectal cancer. Frontiers in Immunology, 11. https://doi.org/10.3389/fimmu.2020.00949
[30]. Huang, W., Ye, D., He, W., He, X., Shi, X., & Gao, Y. (2021). Activated but impaired IFN-γ production of mucosal-associated invariant T cells in patients with hepatocellular carcinoma. Journal for ImmunoTherapy of Cancer, 9(11), e003685. https://doi.org/10.1136/jitc-2021-003685
[31]. Ruf, B., Catania, V. V., Wabitsch, S., Ma, C., Diggs, L. P., Zhang, Q., Heinrich, B., Subramanyam, V., Cui, L. L., Pouzolles, M., Evans, C. N., Chari, R., Sakai, S., Oh, S., Barry, C. E., Barber, D. L., & Greten, T. F. (2021). Activating mucosal-associated invariant T cells induces a broad antitumor response. Cancer Immunology Research, 9(9), 1024-1034. https://doi.org/10.1158/2326-6066.cir-20-0925
Cite this article
Guo,T. (2024). Impact of MR1 on lung cancer in the proliferation of MAIT cells, and production of cytokine and perforin. Theoretical and Natural Science,73,17-27.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Oliver AL. Lung cancer: Epidemiology and screening. Surg Clin North Am (2022) 102(3):335–44. 10.1016/j.suc.2021.12.001
[2]. Lu T, Yang X, Huang Y, Zhao M, Li M, Ma K, et al.. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag Res (2019) 11:943–53. 10.2147/CMAR.S187317
[3]. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20(1):105.
[4]. Quaratino S, Forssmann U, Marschner JP. New approaches in immunotherapy for the treatment of lung cancer. Curr Top Microbiol Immunol (2017) 405:1–31. doi: 10.1007/82_2014_428
[5]. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20(9):1110
[6]. Reantragoon R. et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. The Journal of experimental medicine. 209, 761–774 (2012).
[7]. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al.. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. 10.1182/blood-2010-08-303339
[8]. Yang B, Kang H, Fung A, Zhao H, Wang T, Ma D. The role of interleukin 17 in tumour proliferation, angiogenesis, and metastasis. Mediators Inflamm. (2014) 2014:623759. 10.1155/2014/623759
[9]. Raval A, Puri N, Rath PC, Saxena RK. Cytokine regulation of expression of class I MHC antigens. Exp Mol Med.(1998) 30:1–13. 10.1038/emm.1998.1
[10]. Eun Jeong Won, #1 Jae Kyun Ju, #2 Young-Nan Cho, #3 Hye-Mi Jin, 3 Ki-Jeong Park, 3 Tae-Jong Kim, 3 Yong-Soo Kwon, 4 Hae Jin Kee, 5 Jung-Chul Kim, 2 Seung-Jung Kee, 1 and Yong-Wook Park3Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer2016 Nov 15; 7(46): 76274–76290.
[11]. Ling L, Lin Y, Zheng W, Hong S, Tang X, Zhao P, et al.. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci Rep. (2016) 6:20358. 10.1038/srep20358
[12]. Jorgovanovic, D., Song, M., Wang, L., & Zhang, Y. (2020). Roles of IFN-γ in tumor progression and regression: A review. Biomarker Research, 8(1). https://doi.org/10.1186/s40364-020-00228-x
[13]. Ruf, B., Bruhns, M., Babaei, S., Kedei, N., Ma, L., Revsine, M., Benmebarek, M., Ma, C., Heinrich, B., Subramanyam, V., Qi, J., Wabitsch, S., Green, B. L., Bauer, K. C., Myojin, Y., Greten, L. T., McCallen, J. D., Huang, P., Trehan, R., … Greten, T. F. (2023). Tumor-associated macrophages trigger MAIT cell dysfunction at the HCC invasive margin. Cell, 186(17), 3686-3705.e32. https://doi.org/10.1016/j.cell.2023.07.026
[14]. Zabijak L, Attencourt C, Guignant C, Chatelain D, Marcelo P, Marolleau J-P, et al.. Increased tumor infiltration by mucosal-associated invariant T cells correlates with poor survival in colorectal cancer patients. Cancer Immunol Immunother. (2015) 64:1601–8. 10.1007/s00262-015-1764-7
[15]. Yan, J, Allen, S, McDonald, E, Das, I, Mak, JYW, Liu, L, Fairlie, DP, Meehan, BS, Chen, Z, Corbett, AJ, Varelias, A, Smyth, MJ, Teng, MWL. 2020. MAIT Cells Promote Tumor Initiation, Growth, and Metastases via Tumor MR1. Cancer Discovery. 10:124-141. doi: 10.1158/2159-8290.CD-19-0569. https://www.ncbi.nlm.nih.gov/pubmed/31826876.
[16]. Maimela NR, Liu S, Zhang Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput Struct Biotechnol J. 2018;(xxxx):1–13.
[17]. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
[18]. Zhou, Z., Wang, J., Wang, J., Yang, S., Wang, R., Zhang, G., Li, Z., Shi, R., Wang, Z., & Lu, Q. (2024). Deciphering the tumor immune microenvironment from a multidimensional omics perspective: Insight into next-generation CAR-T cell immunotherapy and beyond. Molecular Cancer, 23(1). https://doi.org/10.1186/s12943-024-02047-2
[19]. Vorwald, V. M., Davis, D. M., Van Gulick, R. J., Torphy, R. J., Borgers, J. S., Klarquist, J., Couts, K. L., Amato, C. M., Cogswell, D. T., Fujita, M., Castleman, M. J., Davis, T., Lozupone, C., Medina, T. M., Robinson, W. A., Gapin, L., McCarter, M. D., & Tobin, R. P. (2022). Circulating CD8<sup>+</sup> mucosal‐associated invariant T cells correlate with improved treatment responses and overall survival in anti‐PD‐1‐treated melanoma patients. Clinical & Translational Immunology, 11(1). https://doi.org/10.1002/cti2.1367
[20]. Sundström, P., Dutta, N., Rodin, W., Hallqvist, A., Raghavan, S., & Quiding Järbrink, M. (2024). Immune checkpoint blockade improves the activation and function of circulating mucosal-associated invariant T (MAIT) cells in patients with non-small cell lung cancer. OncoImmunology, 13(1). https://doi.org/10.1080/2162402x.2024.2312631
[21]. Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16(3):131–144. doi: 10.1038/nrc.2016.14.
[22]. Lopez, J. A., Susanto, O., Jenkins, M. R., Lukoyanova, N., Sutton, V. R., Law, R. H. P., ... & Browne, K. A. (2013). "Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack." Blood, 121(14), 2659-2668.
[23]. Wilke, C. M., Wei, S., Wang, L., Kryczek, I., Kao, J., & Zou, W. (2011). Dual biological effects of the cytokines interleukin-10 and interferon-γ. Cancer Immunology, Immunotherapy, 60(11), 1529-1541. https://doi.org/10.1007/s00262-011-1104-5
[24]. Mocellin S, Panelli M, Wang E, Rossi CR, Pilati P, Nitti D, et al.. IL-10 stimulatory effects on human NK cells explored by gene profile analysis. Genes Immunity. (2004) 5:621–30. 10.1038/sj.gene.6364135
[25]. Park JY, Lee SH, Yoon SR, Park YJ, Jung H, Kim TD, et al.. IL-15-induced IL-10 increases the cytolytic activity of human natural killer cells. Mol Cells. (2011) 32:265–72. 10.1007/s10059-
[26]. Carlini, V., Noonan, D. M., Abdalalem, E., Goletti, D., Sansone, C., Calabrone, L., & Albini, A. (2023). The multifaceted nature of IL-10: Regulation, role in immunological homeostasis and its relevance to cancer, COVID-19 and post-COVID conditions. Frontiers in Immunology, 14. https://doi.org/10.3389/fimmu.2023.1161067
[27]. Shaler, C. R., Tun-Abraham, M. E., Skaro, A. I., Khazaie, K., Corbett, A. J., Mele, T., Hernandez-Alejandro, R., & Haeryfar, S. M. (2017). Mucosa-associated invariant T cells infiltrate hepatic metastases in patients with colorectal carcinoma but are rendered dysfunctional within and adjacent to tumor microenvironment. Cancer Immunology, Immunotherapy, 66(12), 1563-1575. https://doi.org/10.1007/s00262-017-2050-7
[28]. Liu J, Duan Y, Cheng X, et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochemical and Biophysical Research Communications. 2011;407(2):348–354.
[29]. Berzins, S. P., Wallace, M. E., Kannourakis, G., & Kelly, J. (2020). A role for MAIT cells in colorectal cancer. Frontiers in Immunology, 11. https://doi.org/10.3389/fimmu.2020.00949
[30]. Huang, W., Ye, D., He, W., He, X., Shi, X., & Gao, Y. (2021). Activated but impaired IFN-γ production of mucosal-associated invariant T cells in patients with hepatocellular carcinoma. Journal for ImmunoTherapy of Cancer, 9(11), e003685. https://doi.org/10.1136/jitc-2021-003685
[31]. Ruf, B., Catania, V. V., Wabitsch, S., Ma, C., Diggs, L. P., Zhang, Q., Heinrich, B., Subramanyam, V., Cui, L. L., Pouzolles, M., Evans, C. N., Chari, R., Sakai, S., Oh, S., Barry, C. E., Barber, D. L., & Greten, T. F. (2021). Activating mucosal-associated invariant T cells induces a broad antitumor response. Cancer Immunology Research, 9(9), 1024-1034. https://doi.org/10.1158/2326-6066.cir-20-0925









