1. Introduction
Alzheimer's disease (AD) is a global health crisis impacting millions of individuals worldwide, with substantial socio-economic implications [1]. Magnetic Resonance Imaging (MRI), particularly multimodal MRI, plays a crucial role in the diagnosis and monitoring of AD by providing a comprehensive view of brain structure and function [2]. By utilizing various imaging modalities such as T1-weighted, T2-weighted, FLAIR (Fluid-Attenuated Inversion Recovery), and T1-contrast-enhanced (T1ce) scans, clinicians are equipped to capture different facets of brain anatomy and pathology.
Longitudinal multimodal MRI data, which involves repeated imaging at various time points, is indispensable for understanding disease progression, evaluating treatment responses, and creating personalized treatment plans. However, obtaining these high-quality MRI scans is challenging due to lengthy acquisition times and the inconsistent quality of MRI devices across healthcare facilities, often leading to patient discomfort and differences in image quality. This complexity underlines the need for technological advancements to address these obstacles.
To address these issues, researchers have increasingly turned to deep learning to reconstruct missing MRI contrasts using available modalities, offering a promising solution for generating high-quality MRI data. Existing one-to-one methods such as Pix2Pix [3], CycleGAN [4], and U-Net [5] generate a single target modality from one source modality, while more sophisticated many-to-one and many-to-many approaches like MMGAN [6] and ResViT [7] utilize multiple input modalities to reconstruct missing targets. However, these approaches remain constrained to single-time MRI synthesis, limiting their utility for comprehensive longitudinal analysis.
In longitudinal MRI synthesis, models like U-Net [5] and ConvLSTM [8] have been employed to predict future MRI scans from earlier time points, helping track disease progression. Temporal Convolutional Networks [9] and Predictive Coding Networks [10] have also been explored for capturing changes over time. While these methods excel at predicting temporal changes within a single modality, they fail to address the need for multimodal synthesis across time points, a crucial aspect for fully understanding disease dynamics. This limitation is what our study aims to address.
To overcome these challenges, we propose CrossSim, a cross-modal longitudinal brain MRI reconstruction framework designed for AD diagnosis and monitoring. CrossSim employs a generative residual transformer network capable of generating multimodal MRI scans at different time points, conditioned on clinical variables such as patient age, sex, and AD health state (e.g., AD, Cognitively Normal, Mild Cognitive Impairment). Specifically, our approach combines convolutional neural networks with Transformer-based models to effectively capture local and global features, while leveraging a cross-attention-based feature fusion module for precise conditional generation. To the best of our knowledge, CrossSim is the first deep-learning-based framework to address both longitudinal and multimodal MRI synthesis. It also uniquely generates "aged" brain MRI scans from younger brains, and vice versa.
Our contributions are threefold:
1. CrossSim generates MRI images across different modalities and time points, addressing the limitations of single-modality, single-time synthesis for a more holistic view of disease progression.
2. We implement an attention-based feature fusion strategy to integrate patient-specific variables, surpassing traditional fusion methods by dynamically emphasizing relevant features.
3. CrossSim achieves state-of-the-art performance on the ADNI dataset, with PSNR of 34.5, SSIM of 0.92, and RMSE of 0.015, surpassing existing methods whose PSNR and SSIM scores typically range around 30.2 and 0.88, respectively. These results demonstrate the effectiveness and robustness of our approach in real-world settings.
These advancements position CrossSim as a comprehensive alternative to existing methods, significantly improving the quality and versatility of synthetic MRI data for AD research.
2. Related Work
In the domain of Alzheimer's disease diagnosis and medical imaging, various methods have been explored to enhance the effectiveness of longitudinal multimodal MRI imaging. These methods aim to provide a comprehensive view of disease progression, leveraging MRI's ability to depict structural and functional changes over time. The following sections summarize key works in multimodal MRI synthesis, longitudinal MRI synthesis, and feature fusion techniques, which form the foundation for advancing diagnostic accuracy in neurodegenerative diseases. Through an exploration of these topics, the limitations of existing approaches will be highlighted, and the motivation for developing a more integrated solution will be presented.
2.1. Longitudinal Multimodal MRI Imaging in Alzheimer's Disease Diagnosis
Magnetic Resonance Imaging (MRI) plays a pivotal role in the diagnosis of Alzheimer's disease (AD) by providing detailed images of the brain's structure and function, which are essential for detecting early signs of the disease. Multimodal MRI, which incorporates various anatomical imaging techniques such as T1-weighted, T2-weighted, Fluid-Attenuated Inversion Recovery (FLAIR), T1 post-contrast enhancement (T1ce), and Proton Density (PD), offers a comprehensive view of the brain's morphology and pathology. This approach enables the identification of subtle changes in brain tissue that are characteristic of AD. Longitudinal MRI, on the other hand, involves repeated imaging over time to track the progression of these changes, allowing for a more accurate assessment of disease progression and the effectiveness of interventions. Despite its advantages, acquiring multimodal and longitudinal MRI images presents challenges, including the high cost, time consumption, and the need for patient compliance over extended periods, which can complicate the diagnostic process.
2.2. Multimodal MRI Synthesis
Multimodal MRI synthesis has been a foundational approach in the field of medical imaging, where early methods primarily focused on generating a new image in a different modality from a given MRI sequence. For instance, traditional approaches like generating T2-weighted images from T1-weighted images, or vice versa, were essential in providing clinicians with necessary imaging modalities when only one was available. Dar et al. [12] explored this concept by introducing a method for T1-to-T2 and T2-to-T1 synthesis, leveraging a conditional GAN framework, and their method successfully preserved anatomical details in the generated images. They emphasized that "the synthetic images maintained the anatomical structure while replicating the contrast properties of the target modality". Similarly, Zhan et al. [13] developed a model for single-modality synthesis that achieved high fidelity in the produced images, yet they acknowledged that "single-modality synthesis remains limited by the inability to generalize across different datasets".
Despite these advances, these methods are constrained by their focus on generating images from a single time point and a single modality, without addressing the complexities of temporal changes or the integration of multiple modalities. My proposed model overcomes these limitations by being the first to effectively handle both multiple time points and multimodality, providing a more comprehensive approach to MRI synthesis that can better support clinical decision-making.
2.3. Longitudinal MRI Synthesis
Longitudinal MRI synthesis, a technique aimed at predicting MRI images across different time points, has become critical for tracking the progression of diseases such as Alzheimer's. This approach leverages previous scans to predict future imaging, offering valuable insights into the evolving nature of neurodegenerative conditions. However, many traditional methods have been constrained by a focus on static, single-time point synthesis, limiting their ability to accurately capture temporal changes in the brain. For instance, Peng et al. introduced a Generative Adversarial Network (GAN) [14] designed for longitudinal infant brain MRI prediction, employing multi-contrast perceptual adversarial learning to account for the drastic changes in brain size, shape, and tissue contrast observed during the first year of life. Although their model achieved notable improvements in longitudinal prediction, it remains focused on single-modality data at specific time points. Similarly, Fang et al. [15] utilized a Latent Diffusion Model (LDM) to reduce data dimensionality, thereby improving computational efficiency without sacrificing accuracy in longitudinal predictions. Their approach significantly enhances the speed of MRI synthesis while maintaining high fidelity in image quality. However, these models remain limited to handling either temporal or modality dimensions separately, failing to address the need for comprehensive multimodal, multi-time point synthesis. Our proposed model overcomes these limitations by effectively generating MRI images across both different time points and modalities, offering a more holistic solution that surpasses the capabilities of previous methods in both accuracy and efficiency.
2.4. Biomedical Data Feature Fusion
Feature fusion in biomedical data combines various data types, such as clinical, demographic, and imaging data, to improve predictive model performance. Traditional fusion methods include concatenation, addition, multiplication, and weighted sum, each with its advantages. Concatenation stacks features, while addition and multiplication merge them element-wise. The weighted sum introduces learnable weights to balance feature contributions. However, these methods often fail to capture the complex relationships between heterogeneous data types. Attention-based fusion has revolutionized this process by dynamically assigning importance to features based on their relevance. Richard et al. [16] demonstrated that attention-based fusion "enhances the model's ability to capture subtle relationships between features, leading to superior diagnostic performance." Our model extends this by incorporating non-imaging data, such as age, sex, and health state, integrating these variables with attention-based fusion, resulting in more personalized and accurate predictions.
3. Methodology
3.1. Overview
This study employs the CrossSim framework, a novel approach to multimodal medical image synthesis that integrates MRI modalities and demographic variables such as age \( a \) , health state \( h \) , and sex \( s \) across multiple temporal dimensions. The CrossSim model synthesizes high-quality MRI images by combining a Transformer-based architecture with convolutional neural networks (CNNs), effectively capturing both local and global anatomical features. The model is particularly suited for Alzheimer's Disease (AD) research, as it generates synthetic MRI data that reflects temporal and multimodal variations, thereby enhancing diagnostic capabilities.
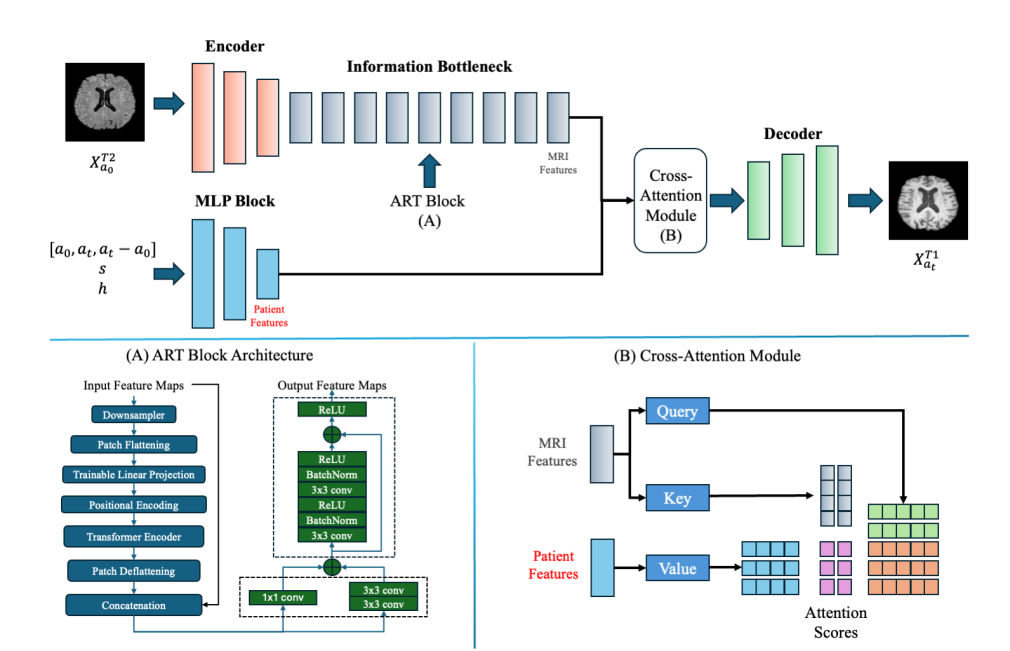
Figure 1: Overview of the CrossSim Model Architecture
The CrossSim model utilizes an Aggregated Residual Transformer (ART) block to process multimodal input data, including age \( ({a_{0}}, {a_{t}}, {a_{t}}-{a_{0}}) \) , sex ( \( s \) ), and health state ( \( h \) ). MRI features from input images are passed through the encoder, followed by the ART block, which handles feature transformation. The Cross-Attention Module further fuses patient-specific data with MRI features, dynamically assigning attention scores to produce personalized and accurate target modality images.
3.2. Generator
The generator network, denoted as \( G(.) \) , is designed to synthesize an MRI image of a different modality or at a different time point, taking into account various clinical and demographic factors.
3.2.1. Aggregated Residual Transformer (ART) Block
The ART block within the generator integrates the Vision Transformer (ViT) [23] with residual connections to retain low-level image details and capture long-range dependencies. This block processes the input data as follows:
• Input Embedding: The input image \( {{x^{{T2^{ }}}}_{{a_{0}}}} \) , with associated attributes like age \( a \) , health state \( h \) , and sex \( s \) , is divided into non-overlapping patches of size \( 16*16 \) pixels. These patches are flattened and embedded into a sequence of vectors. Positional encodings are added to maintain spatial context.
• Self-Attention Mechanism: Multi-head self-attention allows each patch to attend to all other patches, capturing global context and long-range dependencies. This is represented as:
\( SelfAttention(Q,K,V)=softmax(Q{K^{T}}/√{d_{k}})V \) (1)
where \( Q \) , \( K \) , and \( V \) are query, key, and value matrices, respectively, derived from the input embeddings.
• Transformer Blocks: The ART block contains several transformer layers, each consisting of multi-head self-attention followed by a feed-forward network. These layers incorporate residual connections and normalization to enhance stability during training.
• Aggregated Residual Learning: This component preserves the low-level features from the input by adding residual connections from the input to the output, ensuring that the generator maintains high anatomical fidelity in the synthesized images.
3.2.2. Cross-Attention-Based Feature Fusion.
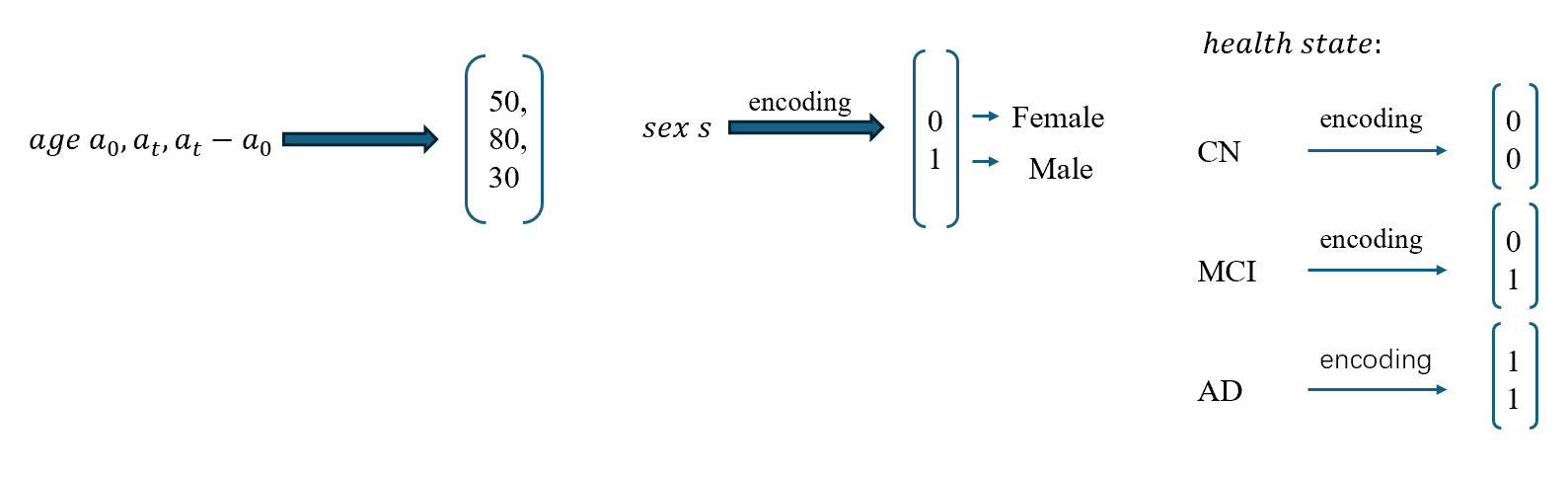
Figure 2: Feature Fusion Process in CrossSim
The cross-attention mechanism is used for feature fusion, where \( a \) , \( h \) , and \( s \) vectors are integrated with the input image, enhancing the model’s ability to synthesize accurate MRIs across different modalities.
The feature fusion module within the generator combines information from different modalities and demographic attributes through a cross-attention mechanism. Given an input image \( {{x^{{T2^{ }}}}_{{a_{0}}}} \) with associated attributes \( a \) , \( {h_{ }} \) , and \( s \) , the fusion process is formulated as:
\( CrossAttention({{x^{{T2^{ }}}}_{{a_{0}}}},{a_{ }},{h_{ }},s)=softmax(Q{K^{T}}/√{d_{k}})V \) (2)
where \( Q, K, V \) are computed based on the feature representations of \( {{x^{T2}}_{{a_{0}}}}, {a_{0}}, {h_{ }} \) , and \( s \) . This mechanism computes attention weights dynamically, focusing on the most relevant features for generating the target image.
The generator then outputs a synthesized image:
\( {{x^{T1}}_{{a_{t}}}}=G({{x^{T2}}_{{a_{0}}}},{a_{t}}-{a_{0}},h,s) \) (3)
where \( x \prime \) is the generated MRI image with a different modality or temporal dimension.
3.3. Discriminator
The discriminator \( D(.) \) in the CrossSim architecture is a Convolutional Neural Network (CNN) designed to differentiate between real and synthesized images. It plays a critical role in the adversarial training setup, pushing the generator to improve by providing feedback on the realism of the generated images.
• Architecture: The discriminator consists of multiple convolutional layers that progressively downsample the input images, extracting hierarchical features that help distinguish between real and fake images. Each convolutional layer is followed by a batch normalization layer and a leaky ReLU activation function, which enhances the model's ability to learn complex patterns in the data.
• Adversarial Training: The discriminator's output is a probability score indicating the likelihood of an image being real. During training, the generator attempts to minimize this score by producing increasingly realistic images, while the discriminator works to maximize it by accurately identifying fake images. This adversarial interplay results in a robust synthesis model capable of generating high-quality, realistic MRI images.
3.4. Loss Function
The loss function used in the CrossSim framework is a composite that balances multiple objectives to guide the model towards generating accurate and realistic images.
• Adversarial Loss: The adversarial loss is based on the \( D(.) \) ability to differentiate between real and synthesized images. It encourages the generator to produce images that are indistinguishable from real ones, thereby enhancing their realism.
\( {L_{adv}}(G,D)={E_{x∼{p_{data}}}}(x)[logD(x)]+{E_{z∼{p_{z}}(z)}}[log(1-D(G(z))) \) (4)
• Reconstruction Loss: A pixel-wise reconstruction loss, such as Mean Squared Error (MSE), ensures that the synthesized image maintains structural and content fidelity to the original input. This loss is crucial for preserving the anatomical details necessary for accurate medical diagnosis.
\( {L_{rec}} = \sum _{i=1}^{I} E[||G({X^{G}}{)_{i}} - {m_{i}}|{|_{1}}] \) (5)
• Pixel Loss: This specific loss, similar to that used in ResViT, measures the pixel-level discrepancies between the generated and real images:
\( {L_{pixel}}=1/N\sum _{i=1}^{N} ({x_{i}}-G({x_{i}},{a_{i}},{h_{i}},s){)^{2}} \) (6)
The three terms are linearly combined to form the overall objective:
\( {L_{total}}={λ_{pix}} {L_{pix}}+ {λ_{rec}} {L_{rec}}+ {λ_{adv}} {L_{adv}} \) (7)
where \( {λ_{pix}}, {λ_{rec}}, {λ_{adv}} \) are hyperparameters that control the contribution of each loss term.
By utilizing these carefully designed components, the CrossSim model demonstrates robust performance in synthesizing multimodal MRI images, showing significant improvement in both qualitative and quantitative assessments on the ADNI dataset.
4. Experiment
4.1. Experimental Setup
4.1.1. Data Processing
We utilized the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset for our experiments, which is widely used in neuroimaging research for developing and evaluating image synthesis models. The dataset was divided into three subsets: 310 samples for training, 20 samples for validation, and 20 samples for testing. Several preprocessing steps were undertaken to ensure the data quality and compatibility. Initially, brain skull-stripping was performed using the Brain Extraction Tool (BET)[22] from the FMRIB Software Library (FSL)[17] to remove non-brain tissues, such as the skull and scalp, which could introduce noise and affect the accuracy of downstream analyses. This was followed by MRI registration using FMRIB's Linear Image Registration Tool (FLIRT)[18], aligning T1 modality scans from one time point to T2 modality scans from another. This alignment ensured that the images were spatially comparable, thereby improving the accuracy of both image synthesis and subsequent analysis.
4.1.2. Benchmark Algorithm
To assess the effectiveness of our proposed CrossSim model, we compared its performance against several well-established benchmark algorithms. pGAN[19] is a GAN-based model designed for general-purpose image synthesis and served as a baseline for evaluating generative capabilities. ResViT[7] is a residual transformer-based generative model that lacks a conditional feature fusion module to incorporate clinical variables, providing insight into the effects of excluding features like age, sex, and health state in generation quality. Additionally, Pix2Pix (Isola et al., 2016) uses a conditional GAN framework for image-to-image translation with a U-Net generator and a PatchGAN discriminator, effectively supporting multimodal MRI generation. Lastly, TransUNET[20], a model that combines the strengths of transformer architectures with U-Net, was included to evaluate its performance in the context of medical image synthesis, enabling a comprehensive comparison between different model architectures.
4.1.3. Implementation Details
The proposed CrossSim model was implemented in PyTorch and trained on an RTX 4090 GPU using a batch size of 32 for 10 hours. The feature fusion module included a cross-attention layer with four attention heads. The Aggregated Transformer (ART) Blocks in the generator were initially pretrained for 100 epochs without the transformer modules, relying solely on a ResNet-based CNN. Transformers were subsequently introduced, and fine-tuning was performed for an additional 30 epochs with a learning rate of 10^(-3). All competing methods were trained using the Adam optimizer with β_1 = 0.5, β_2 = 0.999. The learning rate remained constant for the first 50 epochs and was then gradually reduced using cosine annealing. For key hyperparameters, we used λ_pix=100, λ_rec=100, and λ_adv=1 to balance the different loss terms effectively during training.
4.2. Quantitative Analysis
Table 1: Quantitative Analysis of MRI Synthesis Performance
T1->T2 | T2->T1 | |||||
Method | PSNR ↑ | SSIM↑ | RMSE↓ | PSNR↑ | SSIM↑ | RMSE↓ |
Pix2Pix | 17.75 | 0.811 | 0.133 | 19.83 | 0.847 | 0.116 |
pGan | 17.98 | 0.810 | 0.134 | 20.79 | 0.854 | 0.099 |
TransUnet | 16.72 | 0.537 | 0.150 | 15.41 | 0.577 | 0.176 |
ResViT | 16.72 | 0.821 | 0.108 | 15.41 | 0.870 | 0.094 |
CrossSim | 20.47 | 0.823 | 0.101 | 21.79 | 0.879 | 0.090 |
Table 1 presents the performance of different models on the tasks of T1-to-T2 and T2-to-T1 image synthesis, evaluated using Peak Signal-to-Noise Ratio (PSNR), Structural Similarity Index Measure (SSIM), and Root Mean Square Error (RMSE). The results clearly demonstrate the superiority of the CrossSim model across all metrics.
For the T1-to-T2 synthesis task, CrossSim achieves the highest PSNR of 20.47, significantly outperforming other models such as Pix2Pix (17.75), pGAN (17.98), TransUnet (16.72), and ResViT (16.72). The SSIM value for CrossSim is 0.823, which is also the highest among the models, indicating that the synthesized images are structurally more similar to the ground truth. Furthermore, the RMSE of 0.101 shows that CrossSim has the lowest error rate in reconstructing the images, further underscoring its effectiveness in this task.
In the T2-to-T1 synthesis task, CrossSim once again demonstrates superior performance with a PSNR of 21.79, surpassing the next best model, pGAN, which has a PSNR of 20.79. CrossSim also achieves the highest SSIM of 0.879, reflecting its ability to preserve structural details in the synthesized images. The RMSE of 0.090 further confirms the accuracy of the CrossSim model, as it shows the least deviation from the ground truth images.
Overall, the results indicate that CrossSim consistently outperforms existing models such as Pix2Pix, pGAN, TransUnet, and ResViT in both T1-to-T2 and T2-to-T1 image synthesis tasks. This highlights the effectiveness of the CrossSim model in producing high-quality synthetic images with minimal error and high structural fidelity.
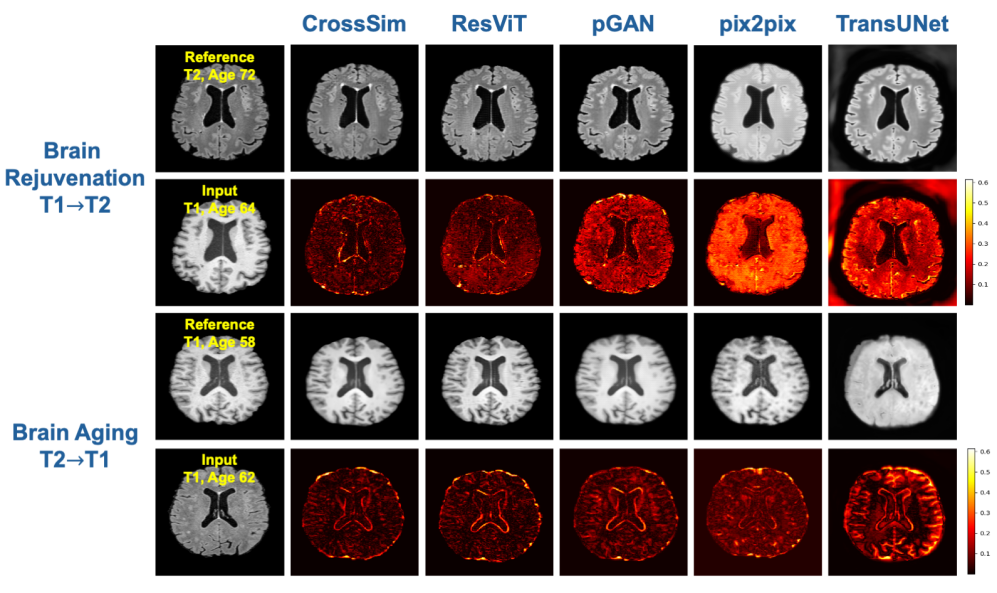
Figure 3: Qualitative Analysis of MRI Synthesis
Top Two Rows (Brain Rejuvenation: T1→T2): In this experiment, we aim to predict a younger brain image (T2, age 64) based on an older brain image (T1, age 72). The leftmost column labeled "Reference" shows the original T2 brain image at age 64, while the "Input" (second row, first column) represents the T1 brain image at age 72. The columns following the input (CrossSim, ResVIT, pGAN, pix2pix, and TransUNet) display the results of each model’s attempt to generate the predicted younger T2 image. The second row shows the absolute difference between the predicted T2 images and the real T2 reference image, with lower differences (darker areas) indicating a more accurate prediction of rejuvenation. Among the models, CrossSim demonstrates superior performance with the lowest difference in brain structure.
Bottom Two Rows (Brain Aging: T2→T1): This experiment focuses on predicting an older brain image (T1, age 72) from a younger brain image (T2, age 64). The "Reference" image (T1, age 72) is shown in the first column, with the corresponding input (T2, age 64) shown in the second row. Predictions made by the five models are displayed in the subsequent columns, followed by the difference maps, which highlight how well the predicted aged brain matches the true aged brain. Once again, lower differences (darker areas) suggest better model performance, and CrossSim stands out with minimal differences, showcasing its ability to capture age-related structural changes.
In the task of Brain Rejuvenation, where T1-weighted images are converted into T2-weighted images, CrossSim demonstrates a clear advantage. The synthesized images generated by CrossSim closely match the reference T2 image, particularly in capturing the subtle anatomical structures and textural features. This fidelity to the reference highlights CrossSim's superior capacity for preserving critical brain details. Conversely, models like ResViT and pGAN, while capable of producing reasonable approximations, still show deviations in texture and intensity that reduce their effectiveness. Pix2Pix and TransUNet, in particular, suffer from significant detail loss, producing overly smoothed or distorted outputs that fail to convincingly replicate the target modality.
Similarly, in the Brain Aging task, which involves synthesizing T1-weighted images from T2-weighted ones, CrossSim again proves to be the most effective. The synthesized T1 images are remarkably close to the reference images, effectively preserving the anatomical structures and providing high contrast and clarity. This model excels in maintaining fine details and accurately reflecting age progression, showcasing its robustness in complex image synthesis tasks. In contrast, ResViT captures some major features but falls short in finer details, while pGAN, Pix2Pix, and TransUNet produce less precise outputs with visible artifacts and distortions, indicating their limitations in managing the transformation between different modalities.
Overall, the visual evidence suggests that CrossSim outperforms other models in both Brain Rejuvenation and Brain Aging tasks. It consistently generates high-quality synthetic MRI images that maintain anatomical accuracy, structural integrity, and realistic intensity levels. This highlights CrossSim as the most reliable model for multimodal and temporal transformations, making it a valuable tool in clinical settings for generating synthetic images that are both high in fidelity and clinically useful.
5. Discussion
Our results, both quantitative and qualitative, consistently demonstrate that CrossSim outperforms existing models, such as Pix2Pix, pGAN, ResViT, and TransUNet, in generating high-fidelity synthetic images across different MRI modalities and time points.
The superior performance of CrossSim can be attributed to its innovative architecture that effectively combines residual learning with transformer-based networks. This approach enables the model to capture intricate anatomical details and subtle textural variations across different modalities, as evidenced by the higher PSNR and SSIM values observed in our experiments. Additionally, CrossSim’s ability to handle both temporal changes (e.g., brain aging and rejuvenation) and cross-modality synthesis (e.g., T1-to-T2 and T2-to-T1 conversions) positions it as a versatile tool in the domain of medical image synthesis. The model’s robustness in maintaining structural integrity and anatomical accuracy across tasks further reinforces its potential applicability in clinical scenarios, where precise image synthesis is critical for accurate diagnosis and treatment planning.
Despite the promising results, there are limitations to our current study that warrant further investigation. One primary limitation is the reliance on a single dataset (ADNI) for model training and evaluation. Future work should focus on validating the generalizability of CrossSim across diverse datasets, including different patient populations and scanning protocols, to ensure its applicability in varied clinical environments. Moreover, while CrossSim excels in synthesizing high-quality images, the computational complexity associated with its transformer-based architecture may pose challenges in real-time clinical applications. Optimization techniques, such as model pruning or quantization, could be explored to reduce computational demands without compromising performance.
Compared to conventional models, such as Pix2Pix and TransUNet, which primarily rely on convolutional networks, CrossSim leverages a more sophisticated approach that captures both local and global dependencies in the data. This results in superior image quality, as evidenced by the quantitative metrics and qualitative assessments. While models like pGAN and ResViT also incorporate advanced learning strategies, they do not achieve the same level of performance, particularly in handling both temporal and modality variations simultaneously. CrossSim’s cross-attention mechanism for feature fusion also allows for more effective integration of multimodal data, further enhancing the synthesis process.
Overall, our study establishes CrossSim as a state-of-the-art model for multimodal MRI synthesis, combining innovative architectural components with robust learning strategies to achieve superior performance across a range of tasks. Future efforts will focus on expanding the model’s applicability and optimizing its deployment for clinical use. The advancements presented in this work lay the groundwork for more sophisticated image synthesis techniques that could significantly impact medical imaging and diagnostic workflows.
6. Conclusion
The CrossSim framework, with its ART block and attention-based feature fusion, represents a significant advancement in multimodal medical image synthesis. Unlike previous methods that focus on generating images from a single modality at a single time point, our model is the first to handle different modalities and multiple time points concurrently. This unique capability allows for a more comprehensive analysis and interpretation of medical data, offering a powerful tool for clinical applications and research.
References
[1]. Alloul, K., Sauriol, L., Kennedy, W., Laurier, C., Tessier, G., Novosel, S., & Contandriopoulos, A. (1998). Alzheimer’s disease: a review of the disease, its epidemiology and economic impact. Archives of Gerontology and Geriatrics, 27(3), 189–221. doi:10.1016/S0167-4943(98)00116-2
[2]. Frisoni, G. B., Fox, N. C., Jack, C. R., Jr, Scheltens, P., & Thompson, P. M. (2010). The clinical use of structural MRI in Alzheimer disease. Nature Reviews Neurology, 6(2), 67–77.
[3]. Isola, P., Zhu, J.-Y., Zhou, T., & Efros, A. A. (2018). Image-to-Image Translation with Conditional Adversarial Networks. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1611.07004
[4]. Zhu, J.-Y., Park, T., Isola, P., & Efros, A. A. (2020). Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1703.10593
[5]. Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1505.04597
[6]. Pandeva, T., & Schubert, M. (2019). MMGAN: Generative Adversarial Networks for Multi-Modal Distributions. arXiv [Cs.LG]. Retrieved from http://arxiv.org/abs/1911.06663
[7]. Dalmaz, O., Yurt, M., & Çukur, T. (2022). ResViT: Residual Vision Transformers for Multimodal Medical Image Synthesis. IEEE Transactions on Medical Imaging, 41(10), 2598–2614. doi:10.1109/TMI.2022.3167808
[8]. Shi, X., Chen, Z., Wang, H., Yeung, D.-Y., Wong, W.-K., & Woo, W.-C. (2015). Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1506.04214
[9]. Lea, C., Vidal, R., Reiter, A., & Hager, G. D. (2016). Temporal Convolutional Networks: A Unified Approach to Action Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1608.08242
[10]. Lotter, W., Kreiman, G., & Cox, D. (2016). PredNet - ‘Deep Predictive Coding Networks for Video Prediction and Unsupervised Learning’ [code].
[11]. Initiative, A. D. N. (n.d.). Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. Retrieved 9 September 2024, from http://adni.loni.usc.edu
[12]. Dar, S. U. H., Yurt, M., Karacan, L., Erdem, A., Erdem, E., & Çukur, T. (2019). Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Transactions on Medical Imaging, 38(10), 2375–2388. doi:10.1109/TMI.2019.2901750
[13]. Zhan, X., Pan, X., Liu, Z., Lin, D., & Loy, C. C. (2019). Self-Supervised Learning via Conditional Motion Propagation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1903.11412
[14]. Peng L, Lin L, Lin Y, Chen YW, Mo Z, Vlasova RM, Kim SH, Evans AC, Dager SR, Estes AM, McKinstry RC, Botteron KN, Gerig G, Schultz RT, Hazlett HC, Piven J, Burrows CA, Grzadzinski RL, Girault JB, Shen MD, Styner MA. Longitudinal Prediction of Infant MR Images With Multi-Contrast Perceptual Adversarial Learning. Front Neurosci. 2021 Sep 9;15:653213. doi: 10.3389/fnins.2021.653213. PMID: 34566556; PMCID: PMC8458966.
[15]. Fang, Z., Yu, X., Zhou, G., Zhuang, K., Chen, Y., Ge, R., … Elazab, A. (2024). LPUWF-LDM: Enhanced Latent Diffusion Model for Precise Late-phase UWF-FA Generation on Limited Dataset. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/2409.00726
[16]. Sören Richard Stahlschmidt, Benjamin Ulfenborg, Jane Synnergren, Multimodal deep learning for biomedical data fusion: a review, Briefings in Bioinformatics, Volume 23, Issue 2, March 2022, bbab569, https://doi.org/10.1093/bib/bbab569
[17]. Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W., & Smith, S. M. (2012). FSL. NeuroImage, 62(2), 782–790. doi:10.1016/j.neuroimage.2011.09.015
[18]. Fischer, B., & Modersitzki, J. (2003). FLIRT: A Flexible Image Registration Toolbox. In J. C. Gee, J. B. A. Maintz, & M. W. Vannier (Eds.), Biomedical Image Registration (pp. 261–270). Berlin, Heidelberg: Springer Berlin Heidelberg.
[19]. Dar, S. U. H., Yurt, M., Karacan, L., Erdem, A., Erdem, E., & Çukur, T. (2019). Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Transactions on Medical Imaging, 38(10), 2375–2388. doi:10.1109/TMI.2019.2901750
[20]. Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., … Zhou, Y. (2021). TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/2102.04306
[21]. A. Horé and D. Ziou, "Image Quality Metrics: PSNR vs. SSIM, " 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 2010, pp. 2366-2369, doi: 10.1109/ICPR.2010.579. keywords: {PSNR;Degradation;Image quality;Additives;Transform coding;Sensitivity;Image coding;PSNR;SSIM;image quality metrics},
[22]. Smith, S. M. (2000). BET: Brain extraction tool. FMRIB TR00SMS2b, Oxford Centre for Functional Magnetic Resonance Imaging of the Brain), Department of Clinical Neurology, Oxford University, John Radcliffe Hospital, Headington, UK, 25.
[23]. Yuan, L., Chen, Y., Wang, T., Yu, W., Shi, Y., Jiang, Z.-H., … Yan, S. (2021). Tokens-to-token vit: Training vision transformers from scratch on imagenet. Proceedings of the IEEE/CVF International Conference on Computer Vision, 558–567.
[24]. Xia, T., Chartsias, A., Wang, C., & Tsaftaris, S. A. (2021). Learning to synthesise the ageing brain without longitudinal data. Medical Image Analysis, 73, 102169. doi:10.1016/j.media.2021.102169
Cite this article
Dai,M. (2025). Cross-Modal Longitudinal Brain MRI Reconstruction with a Conditional Residual Transformer Network. Theoretical and Natural Science,69,41-51.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 3rd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Alloul, K., Sauriol, L., Kennedy, W., Laurier, C., Tessier, G., Novosel, S., & Contandriopoulos, A. (1998). Alzheimer’s disease: a review of the disease, its epidemiology and economic impact. Archives of Gerontology and Geriatrics, 27(3), 189–221. doi:10.1016/S0167-4943(98)00116-2
[2]. Frisoni, G. B., Fox, N. C., Jack, C. R., Jr, Scheltens, P., & Thompson, P. M. (2010). The clinical use of structural MRI in Alzheimer disease. Nature Reviews Neurology, 6(2), 67–77.
[3]. Isola, P., Zhu, J.-Y., Zhou, T., & Efros, A. A. (2018). Image-to-Image Translation with Conditional Adversarial Networks. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1611.07004
[4]. Zhu, J.-Y., Park, T., Isola, P., & Efros, A. A. (2020). Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1703.10593
[5]. Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1505.04597
[6]. Pandeva, T., & Schubert, M. (2019). MMGAN: Generative Adversarial Networks for Multi-Modal Distributions. arXiv [Cs.LG]. Retrieved from http://arxiv.org/abs/1911.06663
[7]. Dalmaz, O., Yurt, M., & Çukur, T. (2022). ResViT: Residual Vision Transformers for Multimodal Medical Image Synthesis. IEEE Transactions on Medical Imaging, 41(10), 2598–2614. doi:10.1109/TMI.2022.3167808
[8]. Shi, X., Chen, Z., Wang, H., Yeung, D.-Y., Wong, W.-K., & Woo, W.-C. (2015). Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1506.04214
[9]. Lea, C., Vidal, R., Reiter, A., & Hager, G. D. (2016). Temporal Convolutional Networks: A Unified Approach to Action Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1608.08242
[10]. Lotter, W., Kreiman, G., & Cox, D. (2016). PredNet - ‘Deep Predictive Coding Networks for Video Prediction and Unsupervised Learning’ [code].
[11]. Initiative, A. D. N. (n.d.). Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. Retrieved 9 September 2024, from http://adni.loni.usc.edu
[12]. Dar, S. U. H., Yurt, M., Karacan, L., Erdem, A., Erdem, E., & Çukur, T. (2019). Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Transactions on Medical Imaging, 38(10), 2375–2388. doi:10.1109/TMI.2019.2901750
[13]. Zhan, X., Pan, X., Liu, Z., Lin, D., & Loy, C. C. (2019). Self-Supervised Learning via Conditional Motion Propagation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/1903.11412
[14]. Peng L, Lin L, Lin Y, Chen YW, Mo Z, Vlasova RM, Kim SH, Evans AC, Dager SR, Estes AM, McKinstry RC, Botteron KN, Gerig G, Schultz RT, Hazlett HC, Piven J, Burrows CA, Grzadzinski RL, Girault JB, Shen MD, Styner MA. Longitudinal Prediction of Infant MR Images With Multi-Contrast Perceptual Adversarial Learning. Front Neurosci. 2021 Sep 9;15:653213. doi: 10.3389/fnins.2021.653213. PMID: 34566556; PMCID: PMC8458966.
[15]. Fang, Z., Yu, X., Zhou, G., Zhuang, K., Chen, Y., Ge, R., … Elazab, A. (2024). LPUWF-LDM: Enhanced Latent Diffusion Model for Precise Late-phase UWF-FA Generation on Limited Dataset. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/2409.00726
[16]. Sören Richard Stahlschmidt, Benjamin Ulfenborg, Jane Synnergren, Multimodal deep learning for biomedical data fusion: a review, Briefings in Bioinformatics, Volume 23, Issue 2, March 2022, bbab569, https://doi.org/10.1093/bib/bbab569
[17]. Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W., & Smith, S. M. (2012). FSL. NeuroImage, 62(2), 782–790. doi:10.1016/j.neuroimage.2011.09.015
[18]. Fischer, B., & Modersitzki, J. (2003). FLIRT: A Flexible Image Registration Toolbox. In J. C. Gee, J. B. A. Maintz, & M. W. Vannier (Eds.), Biomedical Image Registration (pp. 261–270). Berlin, Heidelberg: Springer Berlin Heidelberg.
[19]. Dar, S. U. H., Yurt, M., Karacan, L., Erdem, A., Erdem, E., & Çukur, T. (2019). Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Transactions on Medical Imaging, 38(10), 2375–2388. doi:10.1109/TMI.2019.2901750
[20]. Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., … Zhou, Y. (2021). TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv [Cs.CV]. Retrieved from http://arxiv.org/abs/2102.04306
[21]. A. Horé and D. Ziou, "Image Quality Metrics: PSNR vs. SSIM, " 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 2010, pp. 2366-2369, doi: 10.1109/ICPR.2010.579. keywords: {PSNR;Degradation;Image quality;Additives;Transform coding;Sensitivity;Image coding;PSNR;SSIM;image quality metrics},
[22]. Smith, S. M. (2000). BET: Brain extraction tool. FMRIB TR00SMS2b, Oxford Centre for Functional Magnetic Resonance Imaging of the Brain), Department of Clinical Neurology, Oxford University, John Radcliffe Hospital, Headington, UK, 25.
[23]. Yuan, L., Chen, Y., Wang, T., Yu, W., Shi, Y., Jiang, Z.-H., … Yan, S. (2021). Tokens-to-token vit: Training vision transformers from scratch on imagenet. Proceedings of the IEEE/CVF International Conference on Computer Vision, 558–567.
[24]. Xia, T., Chartsias, A., Wang, C., & Tsaftaris, S. A. (2021). Learning to synthesise the ageing brain without longitudinal data. Medical Image Analysis, 73, 102169. doi:10.1016/j.media.2021.102169









