1. Introduction
With the development of genetic modification using CRISPR, along with improvements in computational and imaging power, we are now witnessing the dawn of a new era in disease diagnosis and personalized genetic susceptibility prediction. The genome editing technique CRISPR/Cas9 has significantly altered biomedical investigation, impacting not only people with genetic diseases but also products and farming methods [1]. A National Institutes of Health (NIH) report states that only 500 of thousands of human diseases have available treatments [2–4]. Genetic changes in the human genome are the root cause of tens of thousands of these diseases. CRISPR technology enables the correction of genetic alteration, which opens up a large number of these diseases as therapeutic interventions [5].
2. Biological mechanism of CRISPR
Prokaryotic adaptive immune system is highly diversified, which includes 33 subtypes, 6 types, and 2 classes of bacterial CRISPR-Cas system [6–7]. All CRISPR/Cas systems still serve the same fundamental purpose despite their extreme diversity, which consists of three primary steps: The process begins with (a) the external DNA pieces being incorporated into the CRISPR array. Next, (b) the adapted CRISPR-RNA (crRNA) is expressed and matures from the acquired spacers [7–8]. Finally, (c) the crRNA that is produced causes interference by recognizing and attaching to a complementary nucleotide sequence, which triggers the Cas nuclease to cleave DNA/RNA [9].
Subsequently, the target gene can be repaired by the machinery of the host cell. There are two primary ways for to carry out the DNA repair: non-homologous end joining (NHEJ) and homology-directed repair (HDR) [10–11].
Delivering the CRISPR components into cells is necessary for therapeutic use in order to accomplish a particular goal.: (a) Cas9/gRNA minimum pair for disruption/mutation of genes; (b) Using a "spare" template DNA and Cas9/gRNA to adjust gene expression; (c) Gene insertion using Cas9/gRNA and a desired gene; and (d) Cas9 and two gRNAs for totally deleting a gene. (Figure1)
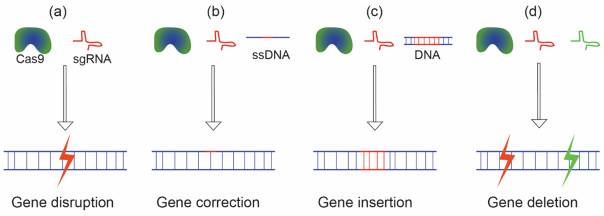
Figure 1. The CRISPR/Cas9 system is injected into cells to accomplish several tasks[12].
In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges paper. (a) Cas9 and sgRNA for knocking out or disrupting genes; (b) A template ssDNA, sgRNA, and Cas9 are used to correct mutations; (c) SgRNA, Cas9, and a template DNA for gene insertion and (d) a pair of sgRNAs and Cas9 to eliminate a gene.
Regardless of the specific components being delivered, the constituents of CRISPR/Cas9 can be introduced into cells using various methods. These delivery methods include: (a) plasmids or viral vectors used in gene-based delivery that contain the Cas9 and sgRNA genes; (b) a synthetic sgRNA and Cas9 mRNA transport with RNA-based and (c) Protein-based delivery using the Cas9 protein and the synthetic sgRNA.
3. CRISPR delivery systems
There are two types of present methods of delivery for prospective CRISPR-based therapies for humans: in vivo and ex vivo [13–16]. And a major challenge to the deficiency of an effective delivery mechanism for CRISPR/Cas9 systems in vivo applications. As a result, investigations have been done to find CRISPR/Cas9 delivery methods that are secure, less immunogenic, biocompatible, and effective. There are two types of delivery mechanisms that can be distinguished: viral and non-viral [17].
3.1. Non-viral delivery of CRISPR/Cas9
3.1.1. Microinjection. One of the most popular methods for introducing CRISPR/Cas9 into cells is microinjection. It entails using a micro needle to inject CRISPR/Cas9 into cells while maintaining microscopic visualization conditions. Because of the difficult technical requirements for manually injecting a single cell and its scalability [18].
3.1.2. Electroporation. One popular technique for both in vitro and ex vivo delivery is electroporation. Through the use of electrical currents, it creates and opens pores in cell membranes, allowing the CRISPR/Cas9 cargo to be internalized. A number of research studies for CRISPR/Cas9 therapeutic delivery have employed ex vivo electroporation techniques. For instance, electroporation has been utilized to treat sickle cell disease (SCD) (Clinical trial number: NCT03745287; NCT03655678) and transfusion-dependent β-thalassemia (TDT)[19]. Even though it was successful in introducing these genome editing instruments into clinical trials, the application of this method is still limited to ex vivo scenarios, and there is a need for further development to enable its in vivo usage without the need to separate cells beforehand for electroporation. Electroporation is a widely utilized technique for both in vitro and ex vivo administration [20].
3.1.3. Hydrodynamic delivery. While microinjection and electroporation can only be used in in vitro or ex vivo settings, hydrodynamic injection has already been utilized to deliver CRISPR/Cas9 in vivo. Using the hydrodynamic injection technique, an important amount of a solution containing the system used by CRISPR/Cas9 is injected into an animal's bloodstream. This results in a rapid increase in hydrodynamic pressure, which momentarily increases endothelial and parenchymal cell permeability and permits the CRISPR/Cas9 payload to enter cells in a variety of tissues, including the muscles, liver, kidneys, heart, and lungs [21]. Unfortunately, because it requires hydrodynamic pressure, which isn't appropriate for in vitro applications, this technically straightforward approach is only available for in vivo applications. Moreover, hydrodynamic delivery causes significant trauma to the body, frequently leading to heart problems, hepatic enlargement, and hypertension [22].
3.1.4. Lipid nanoparticles/liposomes. Lipid nanoparticles have been utilized for a considerable amount of time to transport a variety of molecules into cells, including nucleic acids. Nonetheless, nucleic acids can be relatively easily delivered to cells by encapsulating them within liposomes. Since lipid nanoparticles don't include any viral components, concerns regarding safety and immunogenicity are alleviated or reduced. Two approaches can be used using lipid nanoparticles when utilized as carriers for delivering CRISPR/Cas9 components: either to deliver sgRNA and Cas9 genetic material (plasmid DNA or mRNA), or to deliver Cas9: sgRNA RNP complexes. When it comes to the delivery of Cas9 sgRNA and mRNA, this technique functions similarly to microinjection [23].
3.1.5. Polymer/ DNA nanoclew. With the help of CRISPR/Cas9 systems, polymers can create extremely adaptable molecular complexes that can be functionalized to include a range of elements to tissue targeting / strengthen the cell, endosome escape and cell uptake [24–26]. A novel method for delivering CRISPR/Cas9 components is a DNA nanoclew (NC). The sphere-shaped structure of DNA known as a DNA NC was created by Sun et al. [27]. In a different piece of work, Gu et al. [28] assessed GFP disruption in a model of tumor mouse bearing GFP in the U2OS by delivering Cas9/gRNA using DNA NCs. DNA NCs are DNA nanoparticles designed to partially supplement the gRNA utilized. In order to facilitate endosomal escape, the cationic polymer polyethylenimine has also been applied by the authors to the DNA NCs. According to this study, injecting DNA NCs loaded with Cas9-RNPs intratumorally caused a decrease of 25 percent at the injection site in GFP fluorescence. Before trying to translate the DNA NCs into a clinical setting, more research is necessary to confirm their immunogenic potential.
3.2. Viral delivery of CRISPR/Cas9
3.2.1. Adeno-associated virus (AAV). AAV is a single-stranded DNA virus belonging to the Parvoviridae family and Dependovirus genus. It is frequently used in genetic therapy. There are several compelling reasons why AAV is considered an outstanding means of gene therapy delivery. There is no known link between AAV and any human disease. Additionally, a large variety of recognized serotypes exist that enable infection of numerous cells with various specificities. At least after initial treatment with a serotype, cells can be efficiently infected by the virus itself with little to no innate or adaptive immune reactions or associated damage [29–30]. Lastly, using AAV for gene therapy offers a continuous source of the supplied DNA, in contrast to some other techniques, since the genomic material delivered by the virus can remain exogenous in cells for an indefinite period of time or, with minor modifications, be directly integrated into the host DNA [31]. AAV particles are extremely versatile delivery vehicles due to their capability to be utilized in a wide range of studies, including in vitro, ex vivo, and in vivo applications.
3.2.2. CRISPR-Phage or CR-phage. A recent development in genetics has shown a novel type of modified phage in which the CRISPR/Cas system may be integrated into the bacteriophage genome [32]. The CRISPR-Phage or CR-phage DNA-phage system encapsulates CRISPR-DNA into plasmid constructions. (Figure 2) This novel system may function by adhering to the targeted pathogen through phage, which will allow the CRISPR-conjugated material to be delivered into the pathogen via phage [33–34]. After that, exogenous Cas enzymes penetrate the targeted pathogenic system and reverse genetic alterations, resulting in the faulty genetic makeup that lowers bacterial resistance [35]. This system has an admirable feature in that it can target pathogenic colonies completely with numerous applications of different phage cocktails, which was not achievable before [36]. As a result, the main source of innovation in this system is its integration of both phage and CRISPR-based pathogen knockout specificity, which was absent from earlier research on single system-based therapies [37]. Additionally, this system efficiently regulates the host-pathogen relationship's immuno-dynamics and targeted gene expression, fostering perfect collaboration and enabling successful resistance abrogation, for instance, CR-phages for the treatment of Multi-Drug Resistant (MDR) pathogens. Within the gut microbiota, host-microbiome interactions function in an unusual and synergistic way. Moreover, MDR pathogens typically disrupt the gut microbiota's regular functioning and homeostasis. The delicate balance and homeostasis of the gut are upset by the gradual increase in the pathogen populatio. This dysbiosis leads to alterations in the levels of reactive oxygen species (ROS), disturbances in the intestinal stem cell cycle, and the induction of apoptosis, ultimately impacting intestinal regeneration and homeostasis [38]. In order to fight the MDR pathogens' persistent resistance, pathogen-specific CR-phage development will be an option. In order to completely eradicate bacterial colonies that are populating the gut, these phages will be employed to target specific phage delivery in conjunction with the CRISPR/Cas system against certain resistance genes in pathogens [39].
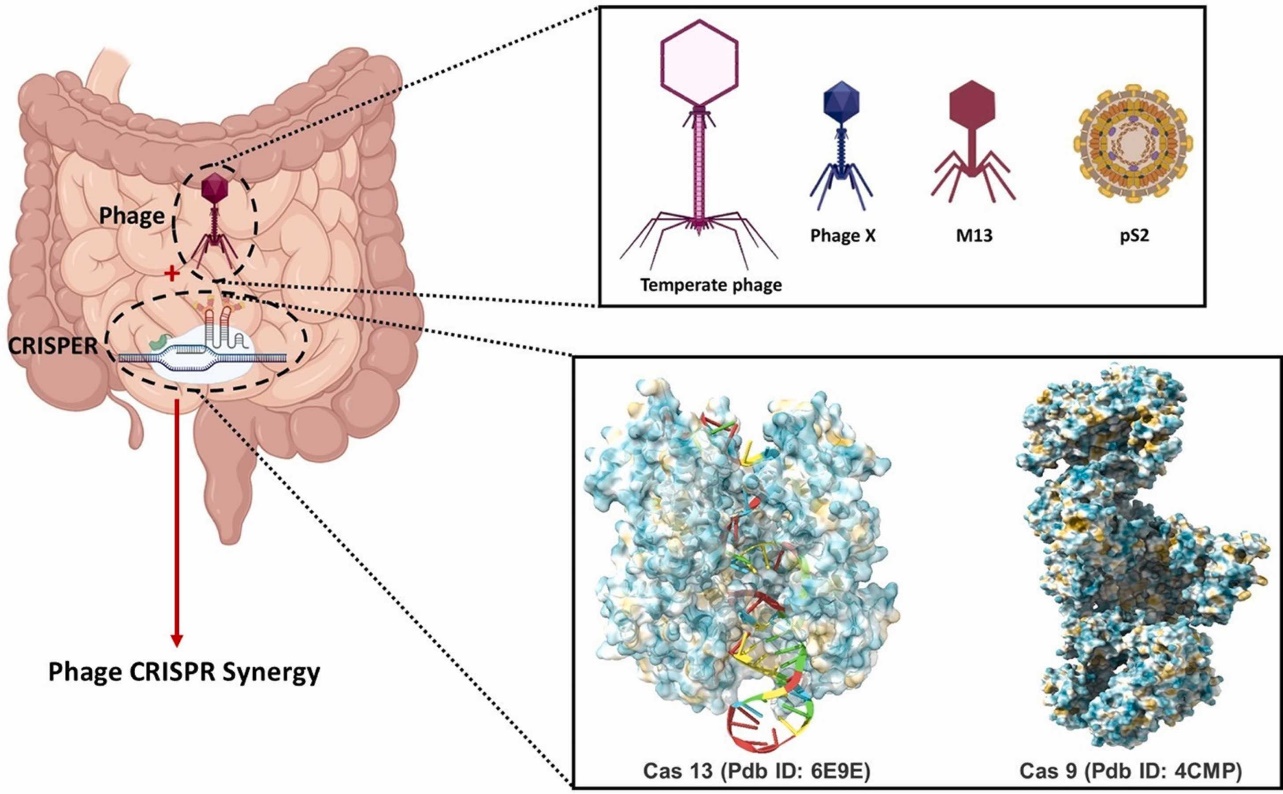
Figure 2. The intestinal niche's phage delivers the CRISPR/Cas system [6].
Phage delivered CRISPR/Cas system to combat multidrug-resistant pathogens in gut microbiome paper). CRISPR/Cas system delivered by phage in intestinal niche. Several phage models can be coupled with specific CRISPR/Cas systems to create the unique modified phage known as CR-Phage. With the ability to introduce CRISPR/Cas systems into specific pathogens directly. A highly specialized instrument called CR-Phage can lessen specific pathogenicity in the intestinal niche. by introducing gene deletions in addition to phage therapy.
4. CHALLENGES
4.1. Packaging challenges
The packaging problem arises in all delivery strategy formats, including RNA, gene, and protein-based delivery. For therapeutic applications, assembling A major problem is incorporating CRISPR components onto a single vector. The maximum size of a cargo gene for gene-based delivery using AAV is approximately 4.7 kilo bases (kbp). Nevertheless, the SpCas9 gene alone is about 4.3 kbp in size. For CRISPR gene delivery using a single AAV vector, it is therefore difficult to insert extra CRISPR components like sgRNA, spare oligonucleotide, or extra genes [40–42]. Additional difficulties arise with protein-based CRISPR delivery because sgRNA (~31 kDa, 5.5 nm hydrodynamic diameter) is negatively charged (~100 PO3− groups), whereas the SpCas9 protein is a substantial protein (160 kDa, ~7.5 nm hydrodynamic diameter) with a net positive surface charge [43]. Packaging these components using supramolecular chemistry could therefore be a significant barrier to developing vehicles for delivery.
4.2. Delivery and editing efficiency
The efficiency of CRISPR/Cas9 in vivo editing is notably inferior to that of in vitro editing. It’s reported that only one out of every 250 edited cells was obtained by hydrodynamic injection of CRISPR components [44]. Another example showed a 20% reduction in GFP fluorescence upon local delivery of Cas9-RNP inserted into a mouse's inner ear. Certain diseases (liver tyrosinemia, muscular dystrophy, etc.) may be sufficiently treated by such a low editing percentage, but other diseases, like cancer, require nearly 100% editing efficiency [45].
4.3. Off-target effect
In order to mitigate the unexpected binding and cleavage effects of CRISPR/Cas nucleases, scientists have used a combination of selective breeding and rational design in order to produce high fidelity Variants of Cas such as SpCas9-HF1 [46], evoCas9 [47], HiFiCas9 [48] and the Cas9 R63A/Q768A variant [49] and direct optimization techniques such as E-Crisp [50–51], CasOFFinder [52], and sgDesigner [53]. These initiatives have produced positive outcomes: according to US Food and Drug Administration (FDA)-grade assays, neither the Intellia sgRNAs nor CRISPR Therapeutics/Vertex, which are currently being used in clinics, contain detectable off-target sites [54–55]. Analysis of base editing results provides an example of How effector domain activity is Cas9-independent, including as deaminases, reverse transcriptases, and transcriptional regulators, can also lead to off-target editing errors. Reducing nucleic acid binding without the aid of Cas is now being accomplished through the use of high fidelity Cas variants and rational deaminase domain engineering [56–60].
4.4. Incidence/efficiency of HDR
In mammalian cells, the frequency of HDR-mediated DNA repair from double strand breaks is generally low. Several strategies, including as the use of tiny molecule inhibitors of NHEJ, have been developed to boost HDR efficiency and reduce NHEJ [61–65], gene silencing [66], cell cycle synchronization [67], and use of cell lines deficient in NHEJ component [68]. Enhancing HDR effectiveness and/or reducing NHEJ is one strategy. It has been demonstrated that single-stranded oligodeoxynucleotide templates can increase HDR efficiency to 60% in human cells for a single-nucleotide substitution [69] , and that cell cycle stage control can favor HDR repair [70–71]. In addition, the donor DNA template can be recruited to the target site using site-specific Cas9-oligonucleotide conjugates [72]. Furthermore, it has been observed that Scr7, one of the most widely used inhibitors, can boost up to 19 folds more HDR from Cas9 editing.
4.5. Immunogenicity
While bacteria are the source of Cas9 and other CRISPR-based genome editing proteins, it is anticipated that these systems may trigger an immunological response from the host. In particular, it is possible for host cells to permanently incorporate the Cas9 gene using CRISPR element delivery. The MHC class I immunological response triggered by the constitutive production of foreign Cas9 protein in the host cell may lead to the host's Cas9-expressing cells being eliminated [73].
5. Conclusion
Gene editing has become more streamlined and precise thanks to the CRISPR/Cas9 system. Nevertheless, their face challenges like off-target effects, s packaging challenges and ethical concern. For this potent gene-editing technique to be implemented responsibly and successfully, these obstacles must be overcome. To guarantee accurate targeting, reduce inadvertent changes, and handle the ethical concerns related to its use, more study are required.
References
[1]. J. Y. Wang and J. A. Doudna, ‘CRISPR technology: A decade of genome editing is only the beginning’, Science, vol. 379, no. 6629, p. eadd8643, Jan. 2023, doi: 10.1126/science.add8643.
[2]. P. D. Hsu, E. S. Lander, and F. Zhang, ‘Development and Applications of CRISPR-Cas9 for Genome Engineering’, Cell, vol. 157, no. 6, pp. 1262–1278, Jun. 2014, doi: 10.1016/j.cell.2014.05.010.
[3]. J. A. Doudna and E. Charpentier, ‘The new frontier of genome engineering with CRISPR-Cas9’, Science, vol. 346, no. 6213, p. 1258096, Nov. 2014, doi: 10.1126/science.1258096.
[4]. D. B. T. Cox, R. J. Platt, and F. Zhang, ‘Therapeutic genome editing: prospects and challenges’, Nat Med, vol. 21, no. 2, pp. 121–131, Feb. 2015, doi: 10.1038/nm.3793.
[5]. ‘National Center for Advancing Translational Sciences’, National Center for Advancing Translational Sciences. Accessed: Sep. 21, 2023. [Online]. Available: https://ncats.nih.gov/
[6]. A. Nath et al., ‘Phage delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiome’, Biomedicine & Pharmacotherapy, vol. 151, p. 113122, Jul. 2022, doi: 10.1016/j.biopha.2022.113122.
[7]. P. Gholizadeh et al., ‘How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance’, IDR, vol. Volume 13, pp. 1111–1121, Apr. 2020, doi: 10.2147/IDR.S247271.
[8]. ‘Evolution and classification of the CRISPR–Cas systems | Nature Reviews Microbiology’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.nature.com/articles/nrmicro2577
[9]. ‘Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials | Nature Biotechnology’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.nature.com/articles/nbt.3043
[10]. ‘CRISPR-Cas systems for genome editing, regulation and targeting - PMC’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4022601/
[11]. F. Wimmer and C. L. Beisel, ‘CRISPR-Cas Systems and the Paradox of Self-Targeting Spacers’, Frontiers in Microbiology, vol. 10, 2020, Accessed: Sep. 22, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03078
[12]. R. Mout, M. Ray, Y.-W. Lee, F. Scaletti, and V. M. Rotello, ‘In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges’, Bioconjug Chem, vol. 28, no. 4, pp. 880–884, Apr. 2017, doi: 10.1021/acs.bioconjchem.7b00057.
[13]. C. A. Lino, J. C. Harper, J. P. Carney, and J. A. Timlin, ‘Delivering CRISPR: a review of the challenges and approaches’, Drug Deliv, vol. 25, no. 1, pp. 1234–1257, Nov. 2018, doi: 10.1080/10717544.2018.1474964.
[14]. S. Tong, B. Moyo, C. M. Lee, K. Leong, and G. Bao, ‘Engineered materials for in vivo delivery of genome-editing machinery’, Nat Rev Mater, vol. 4, pp. 726–737, Nov. 2019, doi: 10.1038/s41578-019-0145-9.
[15]. J. van Haasteren, J. Li, O. J. Scheideler, N. Murthy, and D. V. Schaffer, ‘The delivery challenge: fulfilling the promise of therapeutic genome editing’, Nat Biotechnol, vol. 38, no. 7, pp. 845–855, Jul. 2020, doi: 10.1038/s41587-020-0565-5.
[16]. B. H. Yip, ‘Recent Advances in CRISPR/Cas9 Delivery Strategies’, Biomolecules, vol. 10, no. 6, p. 839, May 2020, doi: 10.3390/biom10060839.
[17]. F. Sinclair, A. A. Begum, C. C. Dai, I. Toth, and P. M. Moyle, ‘Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing’, Drug Deliv Transl Res, vol. 13, no. 5, pp. 1500–1519, May 2023, doi: 10.1007/s13346-023-01320-z.
[18]. L. L. Lesueur, L. M. Mir, and F. M. André, ‘Overcoming the Specific Toxicity of Large Plasmids Electrotransfer in Primary Cells In Vitro’, Mol Ther Nucleic Acids, vol. 5, no. 3, p. e291, Mar. 2016, doi: 10.1038/mtna.2016.4.
[19]. H. Frangoul et al., ‘CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia’, N Engl J Med, vol. 384, no. 3, pp. 252–260, Jan. 2021, doi: 10.1056/NEJMoa2031054.
[20]. J. L. Young and D. A. Dean, ‘Electroporation-Mediated Gene Delivery’, Advances in genetics, vol. 89, p. 49, 2015, doi: 10.1016/bs.adgen.2014.10.003.
[21]. C. A. Lino, J. C. Harper, J. P. Carney, and J. A. Timlin, ‘Delivering CRISPR: a review of the challenges and approaches’, Drug Deliv, vol. 25, no. 1, pp. 1234–1257, Nov. 2018, doi: 10.1080/10717544.2018.1474964.
[22]. B. Bonamassa, L. Hai, and D. Liu, ‘Hydrodynamic Gene Delivery and Its Applications in Pharmaceutical Research’, Pharm Res, vol. 28, no. 4, pp. 694–701, Apr. 2011, doi: 10.1007/s11095-010-0338-9.
[23]. H. Yin et al., ‘Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo’, Nat Biotechnol, vol. 34, no. 3, pp. 328–333, Mar. 2016, doi: 10.1038/nbt.3471.
[24]. A. M. Jhaveri and V. P. Torchilin, ‘Multifunctional polymeric micelles for delivery of drugs and siRNA’, Front. Pharmacol., vol. 5, Apr. 2014, doi: 10.3389/fphar.2014.00077.
[25]. A. Mariano, C. Lubrano, U. Bruno, C. Ausilio, N. B. Dinger, and F. Santoro, ‘Advances in Cell-Conductive Polymer Biointerfaces and Role of the Plasma Membrane’, Chem. Rev., vol. 122, no. 4, pp. 4552–4580, Feb. 2022, doi: 10.1021/acs.chemrev.1c00363.
[26]. L. J. Fox, R. M. Richardson, and W. H. Briscoe, ‘PAMAM dendrimer - cell membrane interactions’, Advances in Colloid and Interface Science, vol. 257, pp. 1–18, Jul. 2018, doi: 10.1016/j.cis.2018.06.005.
[27]. ‘Cocoon-Like Self-Degradable DNA Nanoclew for Anticancer Drug Delivery - PMC’. Accessed: Sep. 23, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4210150/
[28]. W. Sun et al., ‘Efficient Delivery of CRISPR-Cas9 for Genome Editing via Self-Assembled DNA Nanoclews’, Angew Chem Int Ed Engl, vol. 54, no. 41, pp. 12029–12033, Oct. 2015, doi: 10.1002/anie.201506030.
[29]. R. J. Samulski and N. Muzyczka, ‘AAV-Mediated Gene Therapy for Research and Therapeutic Purposes’, Annu. Rev. Virol., vol. 1, no. 1, pp. 427–451, Nov. 2014, doi: 10.1146/annurev-virology-031413-085355.
[30]. S. Daya and K. I. Berns, ‘Gene Therapy Using Adeno-Associated Virus Vectors’, Clin Microbiol Rev, vol. 21, no. 4, pp. 583–593, Oct. 2008, doi: 10.1128/CMR.00008-08.
[31]. D. R. Deyle and D. W. Russell, ‘Adeno-associated virus vector integration’, Curr Opin Mol Ther, vol. 11, no. 4, pp. 442–447, Aug. 2009.
[32]. A. Hatoum-Aslan, ‘Phage Genetic Engineering Using CRISPR–Cas Systems’, Viruses, vol. 10, no. 6, p. 335, Jun. 2018, doi: 10.3390/v10060335.
[33]. J. R. Fagen, D. Collias, A. K. Singh, and C. L. Beisel, ‘Advancing the design and delivery of CRISPR antimicrobials’, Current Opinion in Biomedical Engineering, vol. 4, pp. 57–64, Dec. 2017, doi: 10.1016/j.cobme.2017.10.001.
[34]. A. S. A. Dowah and M. R. J. Clokie, ‘Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria’, Biophys Rev, vol. 10, no. 2, pp. 535–542, Apr. 2018, doi: 10.1007/s12551-017-0382-3.
[35]. D. Palacios Araya, K. L. Palmer, and B. A. Duerkop, ‘CRISPR-based antimicrobials to obstruct antibiotic-resistant and pathogenic bacteria’, PLoS Pathog, vol. 17, no. 7, p. e1009672, Jul. 2021, doi: 10.1371/journal.ppat.1009672.
[36]. F. L. Gordillo Altamirano and J. J. Barr, ‘Phage Therapy in the Postantibiotic Era’, Clin Microbiol Rev, vol. 32, no. 2, pp. e00066-18, Mar. 2019, doi: 10.1128/CMR.00066-18.
[37]. P. Gholizadeh et al., ‘How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance’, IDR, vol. Volume 13, pp. 1111–1121, Apr. 2020, doi: 10.2147/IDR.S247271.
[38]. ‘Reactive Oxygen Species in Modulating Intestinal Stem Cell Dynamics and Function | SpringerLink’. Accessed: Sep. 22, 2023. [Online]. Available: https://link.springer.com/article/10.1007/s12015-022-10377-1
[39]. A. Nath et al., ‘Phage delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiome’, Biomedicine & Pharmacotherapy, vol. 151, p. 113122, Jul. 2022, doi: 10.1016/j.biopha.2022.113122.
[40]. Z. Wu, H. Yang, and P. Colosi, ‘Effect of genome size on AAV vector packaging’, Mol Ther, vol. 18, no. 1, pp. 80–86, Jan. 2010, doi: 10.1038/mt.2009.255.
[41]. B. Dong, H. Nakai, and W. Xiao, ‘Characterization of genome integrity for oversized recombinant AAV vector’, Mol Ther, vol. 18, no. 1, pp. 87–92, Jan. 2010, doi: 10.1038/mt.2009.258.
[42]. E. Senís et al., ‘CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox’, Biotechnol J, vol. 9, no. 11, pp. 1402–1412, Nov. 2014, doi: 10.1002/biot.201400046.
[43]. R. Mout et al., ‘Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing’, ACS Nano, vol. 11, no. 3, pp. 2452–2458, Mar. 2017, doi: 10.1021/acsnano.6b07600.
[44]. H. Yin et al., ‘Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype’, Nat Biotechnol, vol. 32, no. 6, pp. 551–553, Jun. 2014, doi: 10.1038/nbt.2884.
[45]. R. Mout et al., ‘Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing’, ACS Nano, vol. 11, no. 3, pp. 2452–2458, Mar. 2017, doi: 10.1021/acsnano.6b07600.
[46]. B. P. Kleinstiver et al., ‘High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects’, Nature, vol. 529, no. 7587, pp. 490–495, Jan. 2016, doi: 10.1038/nature16526.
[47]. A. Casini et al., ‘A highly specific SpCas9 variant is identified by in vivo screening in yeast’, Nat Biotechnol, vol. 36, no. 3, pp. 265–271, Mar. 2018, doi: 10.1038/nbt.4066.
[48]. C. A. Vakulskas et al., ‘A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells’, Nat Med, vol. 24, no. 8, pp. 1216–1224, Aug. 2018, doi: 10.1038/s41591-018-0137-0.
[49]. M. Bratovič et al., ‘Bridge helix arginines play a critical role in Cas9 sensitivity to mismatches’, Nat Chem Biol, vol. 16, no. 5, pp. 587–595, May 2020, doi: 10.1038/s41589-020-0490-4.
[50]. X.-H. Zhang, L. Y. Tee, X.-G. Wang, Q.-S. Huang, and S.-H. Yang, ‘Off-target Effects in CRISPR/Cas9-mediated Genome Engineering’, Mol Ther Nucleic Acids, vol. 4, no. 11, p. e264, Nov. 2015, doi: 10.1038/mtna.2015.37.
[51]. F. Heigwer, G. Kerr, and M. Boutros, ‘E-CRISP: fast CRISPR target site identification’, Nat Methods, vol. 11, no. 2, pp. 122–123, Feb. 2014, doi: 10.1038/nmeth.2812.
[52]. S. Bae, J. Park, and J.-S. Kim, ‘Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases’, Bioinformatics, vol. 30, no. 10, pp. 1473–1475, May 2014, doi: 10.1093/bioinformatics/btu048.
[53]. K. Hiranniramol, Y. Chen, W. Liu, and X. Wang, ‘Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency’, Bioinformatics, vol. 36, no. 9, pp. 2684–2689, May 2020, doi: 10.1093/bioinformatics/btaa041.
[54]. H. Frangoul et al., ‘CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia’, N Engl J Med, vol. 384, no. 3, pp. 252–260, Jan. 2021, doi: 10.1056/NEJMoa2031054.
[55]. J. D. Gillmore et al., ‘CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis’, N Engl J Med, vol. 385, no. 6, pp. 493–502, Aug. 2021, doi: 10.1056/NEJMoa2107454.
[56]. J. Grünewald et al., ‘Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors’, Nature, vol. 569, no. 7756, pp. 433–437, May 2019, doi: 10.1038/s41586-019-1161-z.
[57]. C. Zhou et al., ‘Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis’, Nature, vol. 571, no. 7764, pp. 275–278, Jul. 2019, doi: 10.1038/s41586-019-1314-0.
[58]. H. A. Rees, C. Wilson, J. L. Doman, and D. R. Liu, ‘Analysis and minimization of cellular RNA editing by DNA adenine base editors’, Sci Adv, vol. 5, no. 5, p. eaax5717, May 2019, doi: 10.1126/sciadv.aax5717.
[59]. J. Grünewald et al., ‘CRISPR DNA base editors with reduced RNA off-target and self-editing activities’, Nat Biotechnol, vol. 37, no. 9, pp. 1041–1048, Sep. 2019, doi: 10.1038/s41587-019-0236-6.
[60]. Y. Yu et al., ‘Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity’, Nat Commun, vol. 11, no. 1, p. 2052, Apr. 2020, doi: 10.1038/s41467-020-15887-5.
[61]. M. Srivastava et al., ‘An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression’, Cell, vol. 151, no. 7, pp. 1474–1487, Dec. 2012, doi: 10.1016/j.cell.2012.11.054.
[62]. A. E. Tomkinson, T. R. L. Howes, and N. E. Wiest, ‘DNA ligases as therapeutic targets’, Transl Cancer Res, vol. 2, no. 3, p. 1219, Jun. 2013.
[63]. F. Robert, M. Barbeau, S. Éthier, J. Dostie, and J. Pelletier, ‘Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing’, Genome Med, vol. 7, no. 1, p. 93, Aug. 2015, doi: 10.1186/s13073-015-0215-6.
[64]. S. V. Vartak and S. C. Raghavan, ‘Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing’, FEBS J, vol. 282, no. 22, pp. 4289–4294, Nov. 2015, doi: 10.1111/febs.13416.
[65]. C. Yu et al., ‘Small molecules enhance CRISPR genome editing in pluripotent stem cells’, Cell Stem Cell, vol. 16, no. 2, pp. 142–147, Feb. 2015, doi: 10.1016/j.stem.2015.01.003.
[66]. V. T. Chu et al., ‘Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells’, Nat Biotechnol, vol. 33, no. 5, pp. 543–548, May 2015, doi: 10.1038/nbt.3198.
[67]. S. Lin, B. T. Staahl, R. K. Alla, and J. A. Doudna, ‘Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery’, Elife, vol. 3, p. e04766, Dec. 2014, doi: 10.7554/eLife.04766.
[68]. D. M. Weinstock and M. Jasin, ‘Alternative pathways for the repair of RAG-induced DNA breaks’, Mol Cell Biol, vol. 26, no. 1, pp. 131–139, Jan. 2006, doi: 10.1128/MCB.26.1.131-139.2006.
[69]. C. D. Richardson, G. J. Ray, M. A. DeWitt, G. L. Curie, and J. E. Corn, ‘Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA’, Nat Biotechnol, vol. 34, no. 3, pp. 339–344, Mar. 2016, doi: 10.1038/nbt.3481.
[70]. T. Gutschner, M. Haemmerle, G. Genovese, G. F. Draetta, and L. Chin, ‘Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair’, Cell Rep, vol. 14, no. 6, pp. 1555–1566, Feb. 2016, doi: 10.1016/j.celrep.2016.01.019.
[71]. A. Lomova et al., ‘Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair’, Stem Cells, vol. 37, no. 2, pp. 284–294, Feb. 2019, doi: 10.1002/stem.2935.
[72]. X. Ling et al., ‘Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates’, Sci Adv, vol. 6, no. 15, p. eaaz0051, Apr. 2020, doi: 10.1126/sciadv.aaz0051.
[73]. J. Neefjes, M. L. M. Jongsma, P. Paul, and O. Bakke, ‘Towards a systems understanding of MHC class I and MHC class II antigen presentation’, Nat Rev Immunol, vol. 11, no. 12, pp. 823–836, Nov. 2011, doi: 10.1038/nri3084.
Cite this article
Zhang,Z. (2024). The approaches and challenges in delivery CRISPR/Cas9. Theoretical and Natural Science,35,92-101.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. J. Y. Wang and J. A. Doudna, ‘CRISPR technology: A decade of genome editing is only the beginning’, Science, vol. 379, no. 6629, p. eadd8643, Jan. 2023, doi: 10.1126/science.add8643.
[2]. P. D. Hsu, E. S. Lander, and F. Zhang, ‘Development and Applications of CRISPR-Cas9 for Genome Engineering’, Cell, vol. 157, no. 6, pp. 1262–1278, Jun. 2014, doi: 10.1016/j.cell.2014.05.010.
[3]. J. A. Doudna and E. Charpentier, ‘The new frontier of genome engineering with CRISPR-Cas9’, Science, vol. 346, no. 6213, p. 1258096, Nov. 2014, doi: 10.1126/science.1258096.
[4]. D. B. T. Cox, R. J. Platt, and F. Zhang, ‘Therapeutic genome editing: prospects and challenges’, Nat Med, vol. 21, no. 2, pp. 121–131, Feb. 2015, doi: 10.1038/nm.3793.
[5]. ‘National Center for Advancing Translational Sciences’, National Center for Advancing Translational Sciences. Accessed: Sep. 21, 2023. [Online]. Available: https://ncats.nih.gov/
[6]. A. Nath et al., ‘Phage delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiome’, Biomedicine & Pharmacotherapy, vol. 151, p. 113122, Jul. 2022, doi: 10.1016/j.biopha.2022.113122.
[7]. P. Gholizadeh et al., ‘How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance’, IDR, vol. Volume 13, pp. 1111–1121, Apr. 2020, doi: 10.2147/IDR.S247271.
[8]. ‘Evolution and classification of the CRISPR–Cas systems | Nature Reviews Microbiology’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.nature.com/articles/nrmicro2577
[9]. ‘Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials | Nature Biotechnology’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.nature.com/articles/nbt.3043
[10]. ‘CRISPR-Cas systems for genome editing, regulation and targeting - PMC’. Accessed: Sep. 22, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4022601/
[11]. F. Wimmer and C. L. Beisel, ‘CRISPR-Cas Systems and the Paradox of Self-Targeting Spacers’, Frontiers in Microbiology, vol. 10, 2020, Accessed: Sep. 22, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03078
[12]. R. Mout, M. Ray, Y.-W. Lee, F. Scaletti, and V. M. Rotello, ‘In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges’, Bioconjug Chem, vol. 28, no. 4, pp. 880–884, Apr. 2017, doi: 10.1021/acs.bioconjchem.7b00057.
[13]. C. A. Lino, J. C. Harper, J. P. Carney, and J. A. Timlin, ‘Delivering CRISPR: a review of the challenges and approaches’, Drug Deliv, vol. 25, no. 1, pp. 1234–1257, Nov. 2018, doi: 10.1080/10717544.2018.1474964.
[14]. S. Tong, B. Moyo, C. M. Lee, K. Leong, and G. Bao, ‘Engineered materials for in vivo delivery of genome-editing machinery’, Nat Rev Mater, vol. 4, pp. 726–737, Nov. 2019, doi: 10.1038/s41578-019-0145-9.
[15]. J. van Haasteren, J. Li, O. J. Scheideler, N. Murthy, and D. V. Schaffer, ‘The delivery challenge: fulfilling the promise of therapeutic genome editing’, Nat Biotechnol, vol. 38, no. 7, pp. 845–855, Jul. 2020, doi: 10.1038/s41587-020-0565-5.
[16]. B. H. Yip, ‘Recent Advances in CRISPR/Cas9 Delivery Strategies’, Biomolecules, vol. 10, no. 6, p. 839, May 2020, doi: 10.3390/biom10060839.
[17]. F. Sinclair, A. A. Begum, C. C. Dai, I. Toth, and P. M. Moyle, ‘Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing’, Drug Deliv Transl Res, vol. 13, no. 5, pp. 1500–1519, May 2023, doi: 10.1007/s13346-023-01320-z.
[18]. L. L. Lesueur, L. M. Mir, and F. M. André, ‘Overcoming the Specific Toxicity of Large Plasmids Electrotransfer in Primary Cells In Vitro’, Mol Ther Nucleic Acids, vol. 5, no. 3, p. e291, Mar. 2016, doi: 10.1038/mtna.2016.4.
[19]. H. Frangoul et al., ‘CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia’, N Engl J Med, vol. 384, no. 3, pp. 252–260, Jan. 2021, doi: 10.1056/NEJMoa2031054.
[20]. J. L. Young and D. A. Dean, ‘Electroporation-Mediated Gene Delivery’, Advances in genetics, vol. 89, p. 49, 2015, doi: 10.1016/bs.adgen.2014.10.003.
[21]. C. A. Lino, J. C. Harper, J. P. Carney, and J. A. Timlin, ‘Delivering CRISPR: a review of the challenges and approaches’, Drug Deliv, vol. 25, no. 1, pp. 1234–1257, Nov. 2018, doi: 10.1080/10717544.2018.1474964.
[22]. B. Bonamassa, L. Hai, and D. Liu, ‘Hydrodynamic Gene Delivery and Its Applications in Pharmaceutical Research’, Pharm Res, vol. 28, no. 4, pp. 694–701, Apr. 2011, doi: 10.1007/s11095-010-0338-9.
[23]. H. Yin et al., ‘Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo’, Nat Biotechnol, vol. 34, no. 3, pp. 328–333, Mar. 2016, doi: 10.1038/nbt.3471.
[24]. A. M. Jhaveri and V. P. Torchilin, ‘Multifunctional polymeric micelles for delivery of drugs and siRNA’, Front. Pharmacol., vol. 5, Apr. 2014, doi: 10.3389/fphar.2014.00077.
[25]. A. Mariano, C. Lubrano, U. Bruno, C. Ausilio, N. B. Dinger, and F. Santoro, ‘Advances in Cell-Conductive Polymer Biointerfaces and Role of the Plasma Membrane’, Chem. Rev., vol. 122, no. 4, pp. 4552–4580, Feb. 2022, doi: 10.1021/acs.chemrev.1c00363.
[26]. L. J. Fox, R. M. Richardson, and W. H. Briscoe, ‘PAMAM dendrimer - cell membrane interactions’, Advances in Colloid and Interface Science, vol. 257, pp. 1–18, Jul. 2018, doi: 10.1016/j.cis.2018.06.005.
[27]. ‘Cocoon-Like Self-Degradable DNA Nanoclew for Anticancer Drug Delivery - PMC’. Accessed: Sep. 23, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4210150/
[28]. W. Sun et al., ‘Efficient Delivery of CRISPR-Cas9 for Genome Editing via Self-Assembled DNA Nanoclews’, Angew Chem Int Ed Engl, vol. 54, no. 41, pp. 12029–12033, Oct. 2015, doi: 10.1002/anie.201506030.
[29]. R. J. Samulski and N. Muzyczka, ‘AAV-Mediated Gene Therapy for Research and Therapeutic Purposes’, Annu. Rev. Virol., vol. 1, no. 1, pp. 427–451, Nov. 2014, doi: 10.1146/annurev-virology-031413-085355.
[30]. S. Daya and K. I. Berns, ‘Gene Therapy Using Adeno-Associated Virus Vectors’, Clin Microbiol Rev, vol. 21, no. 4, pp. 583–593, Oct. 2008, doi: 10.1128/CMR.00008-08.
[31]. D. R. Deyle and D. W. Russell, ‘Adeno-associated virus vector integration’, Curr Opin Mol Ther, vol. 11, no. 4, pp. 442–447, Aug. 2009.
[32]. A. Hatoum-Aslan, ‘Phage Genetic Engineering Using CRISPR–Cas Systems’, Viruses, vol. 10, no. 6, p. 335, Jun. 2018, doi: 10.3390/v10060335.
[33]. J. R. Fagen, D. Collias, A. K. Singh, and C. L. Beisel, ‘Advancing the design and delivery of CRISPR antimicrobials’, Current Opinion in Biomedical Engineering, vol. 4, pp. 57–64, Dec. 2017, doi: 10.1016/j.cobme.2017.10.001.
[34]. A. S. A. Dowah and M. R. J. Clokie, ‘Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria’, Biophys Rev, vol. 10, no. 2, pp. 535–542, Apr. 2018, doi: 10.1007/s12551-017-0382-3.
[35]. D. Palacios Araya, K. L. Palmer, and B. A. Duerkop, ‘CRISPR-based antimicrobials to obstruct antibiotic-resistant and pathogenic bacteria’, PLoS Pathog, vol. 17, no. 7, p. e1009672, Jul. 2021, doi: 10.1371/journal.ppat.1009672.
[36]. F. L. Gordillo Altamirano and J. J. Barr, ‘Phage Therapy in the Postantibiotic Era’, Clin Microbiol Rev, vol. 32, no. 2, pp. e00066-18, Mar. 2019, doi: 10.1128/CMR.00066-18.
[37]. P. Gholizadeh et al., ‘How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance’, IDR, vol. Volume 13, pp. 1111–1121, Apr. 2020, doi: 10.2147/IDR.S247271.
[38]. ‘Reactive Oxygen Species in Modulating Intestinal Stem Cell Dynamics and Function | SpringerLink’. Accessed: Sep. 22, 2023. [Online]. Available: https://link.springer.com/article/10.1007/s12015-022-10377-1
[39]. A. Nath et al., ‘Phage delivered CRISPR-Cas system to combat multidrug-resistant pathogens in gut microbiome’, Biomedicine & Pharmacotherapy, vol. 151, p. 113122, Jul. 2022, doi: 10.1016/j.biopha.2022.113122.
[40]. Z. Wu, H. Yang, and P. Colosi, ‘Effect of genome size on AAV vector packaging’, Mol Ther, vol. 18, no. 1, pp. 80–86, Jan. 2010, doi: 10.1038/mt.2009.255.
[41]. B. Dong, H. Nakai, and W. Xiao, ‘Characterization of genome integrity for oversized recombinant AAV vector’, Mol Ther, vol. 18, no. 1, pp. 87–92, Jan. 2010, doi: 10.1038/mt.2009.258.
[42]. E. Senís et al., ‘CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox’, Biotechnol J, vol. 9, no. 11, pp. 1402–1412, Nov. 2014, doi: 10.1002/biot.201400046.
[43]. R. Mout et al., ‘Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing’, ACS Nano, vol. 11, no. 3, pp. 2452–2458, Mar. 2017, doi: 10.1021/acsnano.6b07600.
[44]. H. Yin et al., ‘Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype’, Nat Biotechnol, vol. 32, no. 6, pp. 551–553, Jun. 2014, doi: 10.1038/nbt.2884.
[45]. R. Mout et al., ‘Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing’, ACS Nano, vol. 11, no. 3, pp. 2452–2458, Mar. 2017, doi: 10.1021/acsnano.6b07600.
[46]. B. P. Kleinstiver et al., ‘High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects’, Nature, vol. 529, no. 7587, pp. 490–495, Jan. 2016, doi: 10.1038/nature16526.
[47]. A. Casini et al., ‘A highly specific SpCas9 variant is identified by in vivo screening in yeast’, Nat Biotechnol, vol. 36, no. 3, pp. 265–271, Mar. 2018, doi: 10.1038/nbt.4066.
[48]. C. A. Vakulskas et al., ‘A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells’, Nat Med, vol. 24, no. 8, pp. 1216–1224, Aug. 2018, doi: 10.1038/s41591-018-0137-0.
[49]. M. Bratovič et al., ‘Bridge helix arginines play a critical role in Cas9 sensitivity to mismatches’, Nat Chem Biol, vol. 16, no. 5, pp. 587–595, May 2020, doi: 10.1038/s41589-020-0490-4.
[50]. X.-H. Zhang, L. Y. Tee, X.-G. Wang, Q.-S. Huang, and S.-H. Yang, ‘Off-target Effects in CRISPR/Cas9-mediated Genome Engineering’, Mol Ther Nucleic Acids, vol. 4, no. 11, p. e264, Nov. 2015, doi: 10.1038/mtna.2015.37.
[51]. F. Heigwer, G. Kerr, and M. Boutros, ‘E-CRISP: fast CRISPR target site identification’, Nat Methods, vol. 11, no. 2, pp. 122–123, Feb. 2014, doi: 10.1038/nmeth.2812.
[52]. S. Bae, J. Park, and J.-S. Kim, ‘Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases’, Bioinformatics, vol. 30, no. 10, pp. 1473–1475, May 2014, doi: 10.1093/bioinformatics/btu048.
[53]. K. Hiranniramol, Y. Chen, W. Liu, and X. Wang, ‘Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency’, Bioinformatics, vol. 36, no. 9, pp. 2684–2689, May 2020, doi: 10.1093/bioinformatics/btaa041.
[54]. H. Frangoul et al., ‘CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia’, N Engl J Med, vol. 384, no. 3, pp. 252–260, Jan. 2021, doi: 10.1056/NEJMoa2031054.
[55]. J. D. Gillmore et al., ‘CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis’, N Engl J Med, vol. 385, no. 6, pp. 493–502, Aug. 2021, doi: 10.1056/NEJMoa2107454.
[56]. J. Grünewald et al., ‘Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors’, Nature, vol. 569, no. 7756, pp. 433–437, May 2019, doi: 10.1038/s41586-019-1161-z.
[57]. C. Zhou et al., ‘Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis’, Nature, vol. 571, no. 7764, pp. 275–278, Jul. 2019, doi: 10.1038/s41586-019-1314-0.
[58]. H. A. Rees, C. Wilson, J. L. Doman, and D. R. Liu, ‘Analysis and minimization of cellular RNA editing by DNA adenine base editors’, Sci Adv, vol. 5, no. 5, p. eaax5717, May 2019, doi: 10.1126/sciadv.aax5717.
[59]. J. Grünewald et al., ‘CRISPR DNA base editors with reduced RNA off-target and self-editing activities’, Nat Biotechnol, vol. 37, no. 9, pp. 1041–1048, Sep. 2019, doi: 10.1038/s41587-019-0236-6.
[60]. Y. Yu et al., ‘Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity’, Nat Commun, vol. 11, no. 1, p. 2052, Apr. 2020, doi: 10.1038/s41467-020-15887-5.
[61]. M. Srivastava et al., ‘An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression’, Cell, vol. 151, no. 7, pp. 1474–1487, Dec. 2012, doi: 10.1016/j.cell.2012.11.054.
[62]. A. E. Tomkinson, T. R. L. Howes, and N. E. Wiest, ‘DNA ligases as therapeutic targets’, Transl Cancer Res, vol. 2, no. 3, p. 1219, Jun. 2013.
[63]. F. Robert, M. Barbeau, S. Éthier, J. Dostie, and J. Pelletier, ‘Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing’, Genome Med, vol. 7, no. 1, p. 93, Aug. 2015, doi: 10.1186/s13073-015-0215-6.
[64]. S. V. Vartak and S. C. Raghavan, ‘Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing’, FEBS J, vol. 282, no. 22, pp. 4289–4294, Nov. 2015, doi: 10.1111/febs.13416.
[65]. C. Yu et al., ‘Small molecules enhance CRISPR genome editing in pluripotent stem cells’, Cell Stem Cell, vol. 16, no. 2, pp. 142–147, Feb. 2015, doi: 10.1016/j.stem.2015.01.003.
[66]. V. T. Chu et al., ‘Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells’, Nat Biotechnol, vol. 33, no. 5, pp. 543–548, May 2015, doi: 10.1038/nbt.3198.
[67]. S. Lin, B. T. Staahl, R. K. Alla, and J. A. Doudna, ‘Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery’, Elife, vol. 3, p. e04766, Dec. 2014, doi: 10.7554/eLife.04766.
[68]. D. M. Weinstock and M. Jasin, ‘Alternative pathways for the repair of RAG-induced DNA breaks’, Mol Cell Biol, vol. 26, no. 1, pp. 131–139, Jan. 2006, doi: 10.1128/MCB.26.1.131-139.2006.
[69]. C. D. Richardson, G. J. Ray, M. A. DeWitt, G. L. Curie, and J. E. Corn, ‘Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA’, Nat Biotechnol, vol. 34, no. 3, pp. 339–344, Mar. 2016, doi: 10.1038/nbt.3481.
[70]. T. Gutschner, M. Haemmerle, G. Genovese, G. F. Draetta, and L. Chin, ‘Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair’, Cell Rep, vol. 14, no. 6, pp. 1555–1566, Feb. 2016, doi: 10.1016/j.celrep.2016.01.019.
[71]. A. Lomova et al., ‘Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair’, Stem Cells, vol. 37, no. 2, pp. 284–294, Feb. 2019, doi: 10.1002/stem.2935.
[72]. X. Ling et al., ‘Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates’, Sci Adv, vol. 6, no. 15, p. eaaz0051, Apr. 2020, doi: 10.1126/sciadv.aaz0051.
[73]. J. Neefjes, M. L. M. Jongsma, P. Paul, and O. Bakke, ‘Towards a systems understanding of MHC class I and MHC class II antigen presentation’, Nat Rev Immunol, vol. 11, no. 12, pp. 823–836, Nov. 2011, doi: 10.1038/nri3084.









