1. Introduction
Antibodies (Abs) are proteins produced by plasma cells that recognize and neutralize foreign molecules (antigens), thereby helping the body defend against non-self intrusions. Antibody drugs leverage the principle of specific antigen–antibody binding to selectively target and bind antigens, thereby exposing the target to the immune system for precise therapeutic intervention. The first antibody drug approved by the U.S. Food and Drug Administration (FDA) was Orthoclone OKT3 in 1986[1]. With advances in antibody production technologies and improved clinical efficacy, a total of 228 antibody drugs had received marketing approval as of December 31, 2024[2]. Antibody drugs have demonstrated significant clinical efficacy in various disease areas, particularly in oncology and autoimmune disorders[3]. Structurally, antibody drugs can be categorized into monoclonal antibodies (mAbs), bispecific antibodies (BsAbs), and antibody–drug conjugates (ADCs), among others[4]. Due to their low toxicity, high potency, and strong specificity, antibody drugs have experienced faster global market growth compared to other biologics. Traditional antibody development relies on screening B cells from immunized animals. Although technologies such as phage display, humanization, and affinity maturation have improved development efficiency, the process remains costly and time-consuming[5].
Artificial intelligence (AI) refers to a set of technologies that enable machines to simulate human abilities such as learning, understanding, problem-solving, and decision-making[6]. With the continuous advancement of AI, it is gradually becoming a core driving force in drug development, serving as a critical tool across key stages including literature retrieval and data curation, target identification and validation, drug molecule design, drug property evaluation, and clinical trial design. On the one hand, AI addresses the challenges posed by multi-source heterogeneous data and complex molecular structures in antibody drug development by enabling efficient standardized processing and in-depth pattern mining, thereby facilitating antibody design and optimization[7]. On the other hand, through algorithms such as ensemble learning and transfer learning, AI significantly enhances the efficiency and accuracy of high-throughput drug screening, reduces reliance on experimental procedures, and lowers R&D costs[8]. As AI technologies evolve rapidly, new-generation models are continuously optimized to replace legacy systems, thereby reshaping the traditional paradigm of antibody drug development.
2. AI-Driven applications across the antibody drug development pipeline
2.1. Target discovery and validation
Target discovery and validation represent the initial and foundational steps of antibody drug development, determining the direction of subsequent antibody design and ultimately influencing clinical outcomes. Traditional approaches rely on transcriptomics and proteomics to identify and validate potential druggable targets through differential expression analysis, gene editing, and protein–protein interaction experiments[9]. In recent years, AI technologies have significantly enhanced the efficiency and accuracy of this process. For instance, PandaOmics, developed by Insilico Medicine, integrates multi-omics data with natural language processing (NLP) models to automatically identify disease-associated targets and evaluate their druggability. This platform has been widely applied in the early stages of drug discovery for diseases such as cancer and fibrosis[10]. During the target validation stage, AI has also demonstrated substantial potential. For example, BpiPred-3.0, a platform developed by Clifford et al. based on protein language models, can accurately predict linear B-cell epitopes. Zhou et al. further developed SEPPA 3.0, which employs logistic regression models to predict spatial and glycoprotein antigen epitopes, thereby significantly improving the efficiency of antibody drug development[11,12].
2.2. Antibody design and optimization
In the antibody design and optimization stage, elucidating the three-dimensional structure of antigen–antibody complexes is essential for identifying key binding sites and guiding functional modifications. Structural information, particularly within the antibody variable regions, helps reveal binding mechanisms and enhances affinity. However, conventional structural analysis techniques—such as X-ray crystallography and cryo-electron microscopy—require high-quality samples and advanced analytical capabilities, and are often time-consuming and costly[13]. The introduction of AI, especially deep learning, has enabled researchers to predict high-accuracy complex structures directly from amino acid sequences. In recent years, tools such as AlphaFold3[14] and Ig-Fold[15] have gradually replaced traditional methods, significantly improving the efficiency of antibody structure modeling. A representative case is the bispecific antibody drug Bizengri (zenocutuzumab-zbco), which targets HER2/HER3 and was approved by the FDA in 2024. Its structure was predicted using AlphaFold3 to inform subsequent optimization (see Figure 1).
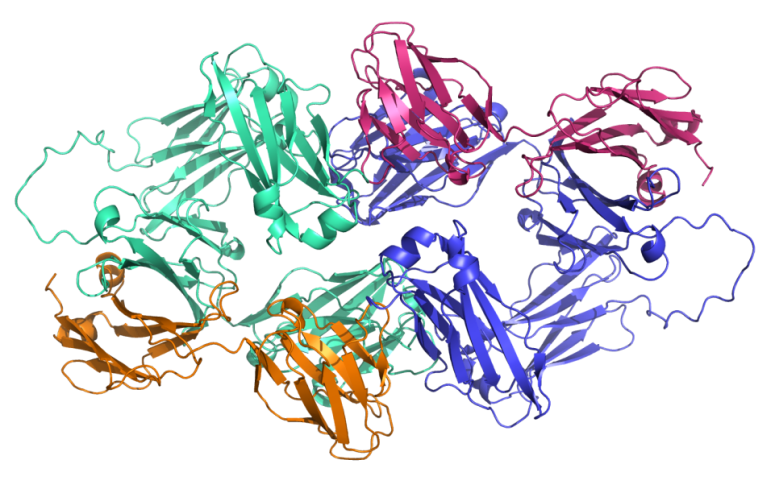
Moreover, AI has shown remarkable advantages in antibody affinity maturation—a process that enhances therapeutic efficacy by optimizing the binding performance between antibodies and antigens[16]. Compared with traditional methods such as FoldX[17] and Rosetta[18], which rely on physical force fields, AI models can guide optimization with greater efficiency and accuracy. The recently released AffinityFlow model demonstrated an affinity improvement rate of up to 93.3%, significantly outperforming conventional approaches[19]. In the area of developability optimization, AI has also introduced novel strategies for antibody refinement. Developability encompasses critical factors such as physicochemical properties, stability, safety, and manufacturability. The CamSol tool, developed by the Oeller group at the University of Cambridge, can predict solubility and identify potential aggregation risks based on the physicochemical properties of amino acids, without requiring known structural information[20]. AI can further be applied to the humanization of antibodies—a process that reduces the immunogenicity of non-human antibodies to enhance drug safety. For instance, BioPhi integrates deep learning with a natural antibody sequence database to enable fully automated, end-to-end humanization design[21], providing strong support for antibody drug development.
2.3. Experimental design and validation
Experimental design and validation are critical steps in determining whether an antibody can effectively bind to its target and exert therapeutic effects. Traditional techniques, such as phage display, are commonly used to screen for antibodies with target specificity and affinity; however, these methods often suffer from long screening cycles and high false-positive rates. To address these limitations, Philpott et al. developed the μCellect platform, which integrates microfluidic technology with machine learning—incorporating both unsupervised and supervised learning algorithms. This system enables the identification of antibodies with picomolar-level affinity in just two rounds of selection, without requiring extensive preliminary experiments, thereby significantly reducing the false-positive rate in prokaryotic expression systems[22].
Moreover, AI-driven virtual experimental design has further reduced reliance on wet-lab experiments, significantly enhancing validation efficiency. A representative case is AU-007, the world’s first antibody drug entirely designed by AI to enter Phase II clinical trials. Biolojic Design simulated the evolutionary process of the immune system and employed AI to screen and optimize potential template antibodies. Based on this, the company conducted antibody engineering by introducing three site-specific mutations, enabling an originally non-binding antibody to specifically target IL-2[23]. AU-007 functions by blocking the interaction between IL-2 and CD25, thereby reducing the activation of regulatory T cells, mitigating IL-2–related side effects, and enhancing antitumor immune responses. This case highlights the tremendous potential of AI in accelerating antibody optimization and clinical translation.
2.4. Preclinical and clinical research
Preclinical and clinical research constitute critical stages for evaluating the safety and efficacy of antibody drugs. Traditionally, the assessment of a drug’s ADMET properties (absorption, distribution, metabolism, excretion, and toxicity) during the preclinical phase relies heavily on extensive wet-lab experiments, which are time-consuming and costly. In recent years, AI models such as ADMETlab 3.0, DeepTox, DeepADMET, pkCSM, and ADMET Predictor have utilized deep learning and graph neural network techniques to efficiently predict pharmacokinetic and toxicological profiles, significantly reducing experimental burden while improving prediction accuracy[24].
During the clinical research phase, antibody drugs often exhibit complex variations in efficacy and safety due to factors such as individual heterogeneity, disease staging, and combination therapies. The integration of real-world data (RWD) can compensate for the limitations of traditional clinical trials by aiding the evaluation of drug performance in diverse populations and guiding personalized treatment[25]. Traditional RWD analysis largely depends on manual processing and rule-based methods, which struggle to handle vast amounts of unstructured information. AI, particularly natural language processing (NLP) and machine learning techniques, can automatically extract critical information from massive, heterogeneous RWD sources, significantly improving both efficiency and analytical depth. For example, in a study of advanced breast cancer, researchers applied quantitative machine learning to analyze digitized pathology images from 154 patients, accurately assessing HER2 expression and predicting the efficacy of the antibody drug trastuzumab deruxtecan, outperforming conventional manual scoring methods[26].
3. Conclusion
AI technology is reshaping the paradigm of antibody drug development, permeating every stage of the process and gradually shifting design approaches from “experience-driven” to “data- and intelligence-driven.” However, numerous challenges remain in practical application. At the technical level, high-quality annotated data are scarce and costly to obtain, limiting model training coverage for long-tail scenarios[27]. Moreover, current AI models, which primarily rely on statistical pattern learning, lack sufficient causal reasoning capabilities, making it difficult to accurately predict key regions such as antibody variable domains. Their “black-box” nature further reduces trustworthiness in clinical settings. At the industrial level, there is a lack of standardized processes bridging AI predictions and wet-lab validation, underscoring the urgent need for professional evaluation protocols to enhance the synergy between computational and experimental efforts. At the ethical level, the patent ownership of AI-generated antibodies remains legally ambiguous, with significant debates over the boundary between algorithmic contribution and human innovation[28]. Furthermore, in AI-assisted personalized therapy, patient biological data privacy protection systems are yet to be fully established, necessitating measures to prevent data misuse. Looking forward, breakthroughs in AI-driven antibody drug development will depend on advancements in multimodal data integration and computational power[29]. Although AI-assisted antibody drug research currently faces both technical and ethical barriers, it is expected that continuous progress in AI will further promote its deep integration across all stages of antibody drug development.
References
[1]. Smith, S. L. (1996). Ten years of orthoclone OKT3 (muromonab-CD3): A review. Journal of Transplant Coordination, 6(3), 109–121. https: //doi.org/10.1177/090591999600600304
[2]. Qian, L., Lin, X., Gao, X., Khan, R. U., Liao, J.-Y., Du, S., Ge, J., Zeng, S., & Yao, S. Q. (2023). The dawn of a new era: Targeting the “undruggables” with antibody-based therapeutics. Chemical Reviews, 123(12), 7782–7853. https: //doi.org/10.1021/acs.chemrev.2c00915
[3]. The Antibody Society. (n.d.). Antibody therapeutics product data. Retrieved April 23, 2025, from https: //www.antibodysociety.org/antibody-therapeutics-product-data/
[4]. Hong, Y., Nam, S.-M., & Moon, A. (2023). Antibody–drug conjugates and bispecific antibodies targeting cancers: Applications of click chemistry. Archives of Pharmacal Research, 46(3), 131–148. https: //doi.org/10.1007/s12272-023-01433-6
[5]. Cheng, J., Liang, T., Xie, X.-Q., Feng, Z., & Meng, L. (2024). A new era of antibody discovery: An in-depth review of AI-driven approaches. Drug Discovery Today, 29(6), 103984. https: //doi.org/10.1016/j.drudis.2024.103984
[6]. Du‐Harpur, X., Watt, F. M., Luscombe, N. M., & Lynch, M. D. (2020). What is AI? Applications of artificial intelligence to dermatology. British Journal of Dermatology, 183(3), 423–430. https: //doi.org/10.1111/bjd.18880
[7]. Norman, R. A., Ambrosetti, F., Bonvin, A. M. J. J., Colwell, L. J., Kelm, S., Kumar, S., & Krawczyk, K. (2020). Computational approaches to therapeutic antibody design: Established methods and emerging trends. Briefings in Bioinformatics, 21(5), 1549–1567. https: //doi.org/10.1093/bib/bbz095
[8]. Dewaker, V., Morya, V. K., Kim, Y. H., Park, S. T., Kim, H. S., & Koh, Y. H. (2025). Revolutionizing oncology: The role of artificial intelligence (AI) as an antibody design, and optimization tools. Biomarker Research, 13(1), 52. https: //doi.org/10.1186/s40364-025-00764-4
[9]. Sleno, L., & Emili, A. (2008). Proteomic methods for drug target discovery. Current Opinion in Chemical Biology, 12(1), 46–54. https: //doi.org/10.1016/j.cbpa.2008.01.022
[10]. Kamya, P., Ozerov, I. V., Pun, F. W., Tretina, K., Fokina, T., Chen, S., Naumov, V., Long, X., Lin, S., Korzinkin, M., Polykovskiy, D., Aliper, A., Ren, F., & Zhavoronkov, A. (2024). PandaOmics: An AI-driven platform for therapeutic target and biomarker discovery. Journal of Chemical Information and Modeling, 64(10), 3961–3969. https: //doi.org/10.1021/acs.jcim.3c01619
[11]. Clifford, J. N., Høie, M. H., Deleuran, S., Peters, B., Nielsen, M., & Marcatili, P. (2022). BepiPred ‐3.0: Improved B‐cell epitope prediction using protein language models. Protein Science, 31(12), e4497. https: //doi.org/10.1002/pro.4497
[12]. Zhou, C., Chen, Z., Zhang, L., Yan, D., Mao, T., Tang, K., Qiu, T., & Cao, Z. (2019). SEPPA 3.0—Enhanced spatial epitope prediction enabling glycoprotein antigens. Nucleic Acids Research, 47(W1), W388–W394. https: //doi.org/10.1093/nar/gkz413
[13]. Bertoline, L. M. F., Lima, A. N., Krieger, J. E., & Teixeira, S. K. (2023). Before and after AlphaFold2: An overview of protein structure prediction. Frontiers in Bioinformatics, 3. https: //doi.org/10.3389/fbinf.2023.1120370
[14]. Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., Ronneberger, O., Willmore, L., Ballard, A. J., Bambrick, J., Bodenstein, S. W., Evans, D. A., Hung, C.-C., O’Neill, M., Reiman, D., Tunyasuvunakool, K., Wu, Z., Žemgulytė, A., Arvaniti, E., … Jumper, J. M. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(8016), 493–500. https: //doi.org/10.1038/s41586-024-07487-w
[15]. Ruffolo, J. A., Chu, L.-S., Mahajan, S. P., & Gray, J. J. (2023). Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. Nature Communications, 14(1), 2389. https: //doi.org/10.1038/s41467-023-38063-x
[16]. Mishra, A. K., & Mariuzza, R. A. (2018). Insights into the structural basis of antibody affinity maturation from next-generation sequencing. Frontiers in Immunology, 9. https: //doi.org/10.3389/fimmu.2018.00117
[17]. Schymkowitz, J., Borg, J., Stricher, F., Nys, R., Rousseau, F., & Serrano, L. (2005). The FoldX web server: An online force field. Nucleic Acids Research, 33(suppl_2), W382–W388. https: //doi.org/10.1093/nar/gki387
[18]. Das, R., & Baker, D. (2008). Macromolecular modeling with Rosetta. Annual Review of Biochemistry, 77, 363–382. https: //doi.org/10.1146/annurev.biochem.77.062906.171838
[19]. Chen, C., Herpoldt, K.-L., Zhao, C., Wang, Z., Collins, M., Shang, S., & Benson, R. (2025). AffinityFlow: Guided flows for antibody affinity maturation (No. arXiv: 2502.10365). arXiv. https: //doi.org/10.48550/arXiv.2502.10365
[20]. Oeller, M., Kang, R., Bell, R., Ausserwöger, H., Sormanni, P., & Vendruscolo, M. (2023). Sequence-based prediction of pH-dependent protein solubility using CamSol. Briefings in Bioinformatics, 24(2), bbad004. https: //doi.org/10.1093/bib/bbad004
[21]. Prihoda, D., Maamary, J., Waight, A., Juan, V., Fayadat-Dilman, L., Svozil, D., & Bitton, D. A. (2022). BioPhi: A platform for antibody design, humanization, and humanness evaluation based on natural antibody repertoires and deep learning. mAbs, 14(1), 2020203. https: //doi.org/10.1080/19420862.2021.2020203
[22]. Philpott, D. N., Gomis, S., Wang, H., Atwal, R., Kelil, A., Sack, T., Morningstar, B., Burnie, C., Sargent, E. H., Angers, S., Sidhu, S., & Kelley, S. O. (2022). Rapid on-cell selection of high-performance human antibodies. ACS Central Science, 8(1), 102–109. https: //doi.org/10.1021/acscentsci.1c01205
[23]. Mullard, A. (2021). Restoring IL-2 to its cancer immunotherapy glory. Nature Reviews Drug Discovery, 20(3), 163–166.
[24]. Huang, D., Yang, M., Wen, X., Xia, S., & Yuan, B. (2024). AI-driven drug discovery: Accelerating the development of novel therapeutics in biopharmaceuticals. Journal of Knowledge Learning and Science Technology, 3(3), Article 3. https: //doi.org/10.60087/jklst.vol3.n3.p.206-224
[25]. Saesen, R., Lacombe, D., & Huys, I. (2023). Real-world data in oncology: A questionnaire-based analysis of the academic research landscape examining the policies and experiences of the cancer cooperative groups. ESMO Open, 8(2), 100878. https: //doi.org/10.1016/j.esmoop.2023.100878
[26]. Modi, S., Glass, B., Prakash, A., Taylor-Weiner, A., Elliott, H., Wapinski, I., Sugihara, M., Saito, K., Kerner, J. K., Phillips, R., Shibutani, T., Honda, K., Khosla, A., Beck, A. H., & Cogswell, J. (2020). 286P Artificial intelligence analysis of advanced breast cancer patients from a phase I trial of trastuzumab deruxtecan (T-DxD): HER2 and histopathology features as predictors of clinical benefit. Annals of Oncology, 31(suppl_4), S355–S356. https: //doi.org/10.1016/j.annonc.2020.08.388
[27]. van der Wal, D., Jhun, I., Laklouk, I., Nirschl, J., Richer, L., Rojansky, R., Theparee, T., Wheeler, J., Sander, J., Feng, F., Mohamad, O., Savarese, S., Socher, R., & Esteva, A. (2021). Biological data annotation via a human-augmenting AI-based labeling system. NPJ Digital Medicine, 4(1), 1–7. https: //doi.org/10.1038/s41746-021-00520-6
[28]. Teli, J. S., Rai, A., & Lin, Y.-K. (2024). Abnormal returns to artificial intelligence patent infringement litigations. Journal of Management Information Systems, 41(2), 422–452. https: //doi.org/10.1080/07421222.2024.2340826
[29]. Dentamaro, V., Impedovo, D., Musti, L., Pirlo, G., & Taurisano, P. (2024). Enhancing early Parkinson’s disease detection through multimodal deep learning and explainable AI: Insights from the PPMI database. Scientific Reports, 14(1), 20941. https: //doi.org/10.1038/s41598-024-70165-4
Cite this article
Xu,N. (2025). Research Progress on Antibody Drug Development under the Guidance of Artificial Intelligence. Theoretical and Natural Science,126,10-15.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Smith, S. L. (1996). Ten years of orthoclone OKT3 (muromonab-CD3): A review. Journal of Transplant Coordination, 6(3), 109–121. https: //doi.org/10.1177/090591999600600304
[2]. Qian, L., Lin, X., Gao, X., Khan, R. U., Liao, J.-Y., Du, S., Ge, J., Zeng, S., & Yao, S. Q. (2023). The dawn of a new era: Targeting the “undruggables” with antibody-based therapeutics. Chemical Reviews, 123(12), 7782–7853. https: //doi.org/10.1021/acs.chemrev.2c00915
[3]. The Antibody Society. (n.d.). Antibody therapeutics product data. Retrieved April 23, 2025, from https: //www.antibodysociety.org/antibody-therapeutics-product-data/
[4]. Hong, Y., Nam, S.-M., & Moon, A. (2023). Antibody–drug conjugates and bispecific antibodies targeting cancers: Applications of click chemistry. Archives of Pharmacal Research, 46(3), 131–148. https: //doi.org/10.1007/s12272-023-01433-6
[5]. Cheng, J., Liang, T., Xie, X.-Q., Feng, Z., & Meng, L. (2024). A new era of antibody discovery: An in-depth review of AI-driven approaches. Drug Discovery Today, 29(6), 103984. https: //doi.org/10.1016/j.drudis.2024.103984
[6]. Du‐Harpur, X., Watt, F. M., Luscombe, N. M., & Lynch, M. D. (2020). What is AI? Applications of artificial intelligence to dermatology. British Journal of Dermatology, 183(3), 423–430. https: //doi.org/10.1111/bjd.18880
[7]. Norman, R. A., Ambrosetti, F., Bonvin, A. M. J. J., Colwell, L. J., Kelm, S., Kumar, S., & Krawczyk, K. (2020). Computational approaches to therapeutic antibody design: Established methods and emerging trends. Briefings in Bioinformatics, 21(5), 1549–1567. https: //doi.org/10.1093/bib/bbz095
[8]. Dewaker, V., Morya, V. K., Kim, Y. H., Park, S. T., Kim, H. S., & Koh, Y. H. (2025). Revolutionizing oncology: The role of artificial intelligence (AI) as an antibody design, and optimization tools. Biomarker Research, 13(1), 52. https: //doi.org/10.1186/s40364-025-00764-4
[9]. Sleno, L., & Emili, A. (2008). Proteomic methods for drug target discovery. Current Opinion in Chemical Biology, 12(1), 46–54. https: //doi.org/10.1016/j.cbpa.2008.01.022
[10]. Kamya, P., Ozerov, I. V., Pun, F. W., Tretina, K., Fokina, T., Chen, S., Naumov, V., Long, X., Lin, S., Korzinkin, M., Polykovskiy, D., Aliper, A., Ren, F., & Zhavoronkov, A. (2024). PandaOmics: An AI-driven platform for therapeutic target and biomarker discovery. Journal of Chemical Information and Modeling, 64(10), 3961–3969. https: //doi.org/10.1021/acs.jcim.3c01619
[11]. Clifford, J. N., Høie, M. H., Deleuran, S., Peters, B., Nielsen, M., & Marcatili, P. (2022). BepiPred ‐3.0: Improved B‐cell epitope prediction using protein language models. Protein Science, 31(12), e4497. https: //doi.org/10.1002/pro.4497
[12]. Zhou, C., Chen, Z., Zhang, L., Yan, D., Mao, T., Tang, K., Qiu, T., & Cao, Z. (2019). SEPPA 3.0—Enhanced spatial epitope prediction enabling glycoprotein antigens. Nucleic Acids Research, 47(W1), W388–W394. https: //doi.org/10.1093/nar/gkz413
[13]. Bertoline, L. M. F., Lima, A. N., Krieger, J. E., & Teixeira, S. K. (2023). Before and after AlphaFold2: An overview of protein structure prediction. Frontiers in Bioinformatics, 3. https: //doi.org/10.3389/fbinf.2023.1120370
[14]. Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., Ronneberger, O., Willmore, L., Ballard, A. J., Bambrick, J., Bodenstein, S. W., Evans, D. A., Hung, C.-C., O’Neill, M., Reiman, D., Tunyasuvunakool, K., Wu, Z., Žemgulytė, A., Arvaniti, E., … Jumper, J. M. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(8016), 493–500. https: //doi.org/10.1038/s41586-024-07487-w
[15]. Ruffolo, J. A., Chu, L.-S., Mahajan, S. P., & Gray, J. J. (2023). Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. Nature Communications, 14(1), 2389. https: //doi.org/10.1038/s41467-023-38063-x
[16]. Mishra, A. K., & Mariuzza, R. A. (2018). Insights into the structural basis of antibody affinity maturation from next-generation sequencing. Frontiers in Immunology, 9. https: //doi.org/10.3389/fimmu.2018.00117
[17]. Schymkowitz, J., Borg, J., Stricher, F., Nys, R., Rousseau, F., & Serrano, L. (2005). The FoldX web server: An online force field. Nucleic Acids Research, 33(suppl_2), W382–W388. https: //doi.org/10.1093/nar/gki387
[18]. Das, R., & Baker, D. (2008). Macromolecular modeling with Rosetta. Annual Review of Biochemistry, 77, 363–382. https: //doi.org/10.1146/annurev.biochem.77.062906.171838
[19]. Chen, C., Herpoldt, K.-L., Zhao, C., Wang, Z., Collins, M., Shang, S., & Benson, R. (2025). AffinityFlow: Guided flows for antibody affinity maturation (No. arXiv: 2502.10365). arXiv. https: //doi.org/10.48550/arXiv.2502.10365
[20]. Oeller, M., Kang, R., Bell, R., Ausserwöger, H., Sormanni, P., & Vendruscolo, M. (2023). Sequence-based prediction of pH-dependent protein solubility using CamSol. Briefings in Bioinformatics, 24(2), bbad004. https: //doi.org/10.1093/bib/bbad004
[21]. Prihoda, D., Maamary, J., Waight, A., Juan, V., Fayadat-Dilman, L., Svozil, D., & Bitton, D. A. (2022). BioPhi: A platform for antibody design, humanization, and humanness evaluation based on natural antibody repertoires and deep learning. mAbs, 14(1), 2020203. https: //doi.org/10.1080/19420862.2021.2020203
[22]. Philpott, D. N., Gomis, S., Wang, H., Atwal, R., Kelil, A., Sack, T., Morningstar, B., Burnie, C., Sargent, E. H., Angers, S., Sidhu, S., & Kelley, S. O. (2022). Rapid on-cell selection of high-performance human antibodies. ACS Central Science, 8(1), 102–109. https: //doi.org/10.1021/acscentsci.1c01205
[23]. Mullard, A. (2021). Restoring IL-2 to its cancer immunotherapy glory. Nature Reviews Drug Discovery, 20(3), 163–166.
[24]. Huang, D., Yang, M., Wen, X., Xia, S., & Yuan, B. (2024). AI-driven drug discovery: Accelerating the development of novel therapeutics in biopharmaceuticals. Journal of Knowledge Learning and Science Technology, 3(3), Article 3. https: //doi.org/10.60087/jklst.vol3.n3.p.206-224
[25]. Saesen, R., Lacombe, D., & Huys, I. (2023). Real-world data in oncology: A questionnaire-based analysis of the academic research landscape examining the policies and experiences of the cancer cooperative groups. ESMO Open, 8(2), 100878. https: //doi.org/10.1016/j.esmoop.2023.100878
[26]. Modi, S., Glass, B., Prakash, A., Taylor-Weiner, A., Elliott, H., Wapinski, I., Sugihara, M., Saito, K., Kerner, J. K., Phillips, R., Shibutani, T., Honda, K., Khosla, A., Beck, A. H., & Cogswell, J. (2020). 286P Artificial intelligence analysis of advanced breast cancer patients from a phase I trial of trastuzumab deruxtecan (T-DxD): HER2 and histopathology features as predictors of clinical benefit. Annals of Oncology, 31(suppl_4), S355–S356. https: //doi.org/10.1016/j.annonc.2020.08.388
[27]. van der Wal, D., Jhun, I., Laklouk, I., Nirschl, J., Richer, L., Rojansky, R., Theparee, T., Wheeler, J., Sander, J., Feng, F., Mohamad, O., Savarese, S., Socher, R., & Esteva, A. (2021). Biological data annotation via a human-augmenting AI-based labeling system. NPJ Digital Medicine, 4(1), 1–7. https: //doi.org/10.1038/s41746-021-00520-6
[28]. Teli, J. S., Rai, A., & Lin, Y.-K. (2024). Abnormal returns to artificial intelligence patent infringement litigations. Journal of Management Information Systems, 41(2), 422–452. https: //doi.org/10.1080/07421222.2024.2340826
[29]. Dentamaro, V., Impedovo, D., Musti, L., Pirlo, G., & Taurisano, P. (2024). Enhancing early Parkinson’s disease detection through multimodal deep learning and explainable AI: Insights from the PPMI database. Scientific Reports, 14(1), 20941. https: //doi.org/10.1038/s41598-024-70165-4









