1. Introduction
As the global population ages, there is a growing demand for interventions that can improve the quality of life in older adults. Over the past century (1920–2020), the growth rate of Americans 65 and older was almost five times higher than that of the total population, as reported by the 2020 Census. By 2020, this age group had reached 55.8 million, making up 16.8% of the U.S. population. (US census) There are many diseases related to Aging, such as Alzheimer’s Disease and Cancer [1]. Thus, Anti-aging research aims to find ways to extend not just lifespan but healthspan—the period during which individuals remain healthy and active, to delay the process of aging-related diseases [2]. Lifespan is regulated by genes: There are some relations between parental lifespan and the offspring's life span [3]. IGF-1, mTOR, AMPK, and APOE are the well-known pathways that regulate the gene in humans, mice, rats, and C. elegans [4]. For example, one of the most well-known pathways that regulate aging in C. elegans is the insulin/IGF-1 signaling (IIS) pathway [3]. Mutations in the gene daf-2, which encodes the insulin/IGF-1 receptor, can significantly extend lifespan by 200% [5]. When daf-2 is downregulated, it leads to activation of the transcription factor DAF-16, which is involved in stress resistance, metabolism, and longevity. The suppression of insulin/IGF-1 signaling mimics a low-nutrient environment, promoting longevity [3]. Some ingredients can extend the lifespan of model organisms by regulating genes, such as the discovery of nematodes [6]. However, the overall regulatory mechanism of aging genes is unclear, which may be the main reason why there are no proven anti-aging drugs on the market. Some compounds may show promise in extending lifespan in the short term in humans [7], but their effects over longer periods and their safety profiles need more thorough investigation. These processes usually take a long time. Also, there might be some ethical and social considerations. In this experiment, we used C. elegans for these advantages: The identified gene map, high similarity to the human genome, easy maintenance and manipulation, small size and large brood size, etc [8]. It allows the study related to genes that are homologous to human genes and favors transgenic expression of human disease-related proteins [9].
This study tested several potential drugs in mammals, including rapamycin, metformin, royal jelly, and rilmenidine, to identify their effect on C. elegans lifespan as well as the mechanisms on genetics.
2. Method
2.1. Strains and growth conditions
C. elegans strain N2 (wild type) was obtained from the C. elegans Genetic Center, CGC (University of Minnesota, Minneapolis, MN, USA) and was maintained at 22.5 ℃ on solid nematode growth medium (NGM), seeded with live E. coli (OP50) as a food source [10] (Figure 1).
2.2. Preparation of medicine
The candidate drugs including rapamycin (50mM), metformin (100 μM), royal jelly (10 μg/ml), and rilmenidine (200 μM) were added to the OP50 bacteria on the standard NGM plates (Table 1).
|
Group |
Medicine |
Work Concentration |
|
1 |
Control |
0 |
|
2 |
Metformin |
50 mM |
|
3 |
Rapamycin |
100 μM |
|
4 |
Royal jelly |
10 μg/ml |
|
5 |
rilmenidine |
200 μM |
2.3. Lifespan assay
The lifespan assay was modified slightly according to the methods previously described [11]. The worms were synchronized to eggs before the experiments. When eggs reached L4 stages, approximately 40 worms were transferred onto new NGM dishes with or without drugs and transferred every day. Worms that were missing, attaching to walls or wormbags were censored.
2.4. Pumping rate assay
The pumping rate phenotype was observed as previously described [12]. Optimize the nematode pumping rate recording by observing the 1st, 5th, 10th, 15th, 20th, 25th, 27th, and 30th days post-drug treatment. For each group, randomly select 10 nematodes to be counted on the OP50 lawn; avoid blank plates. Counting can be done directly on the plate, eliminating the need for transfer. If fewer than 10 nematodes are present on the RNAi plate, count all of them. Measure each nematode for 20 seconds, recording the number of pharynx pumps during this interval (one complete back-and-forth movement counts as one pump). Repeat the counting for each group three times, and document all results. Ensure that the entire experiment is conducted on a single day.
2.5. Expression analysis by quantitative Real-Time RCR
Gene expression analysis was conducted using an adapted qRT-PCR protocol. Synchronized worm populations, both treated and untreated with experimental compounds, were assessed for changes in expression levels of key metabolic and longevity-associated genes (daf-2, age-1, mecr-1, rml-1, idha-1, aco-2, idh-1, cox-5B, and cox-4), with results presented in Table 2. RNA extraction was performed using FastPure® Cell/Tissue Total RNA Isolation Kit V2 (Vazyme, China), followed by cDNA synthesis with the HiScript 1st strand cDNA synthesis kit (Vazyme, China). The housekeeping gene tba-1 served as the normalization control, with all primer sequences detailed in the methods section.
|
Gene name |
Forward Primer |
Reversed Primer |
|
daf-2 |
CACAGATTTGTGATGGTATGGCGTAC |
CGACGTTCCGAATCACTCTGAAC |
|
age-1 |
GTTGTTCGCCGACAATCACTAGTC |
CTTGTCGTAGTTCATCTCTCCAATCG |
|
mecr-1 |
GAATCCTCCCACAGCTTATCGTATG |
CAAGCTCCTGGTACATCTCATGC |
|
rml-1 |
CCAGACTGATCTCGTTTCGTG |
GCTTCGAGTGATTTGACGATTGG |
|
idha-1 |
GTCTACCGTCGGACAATCCATCAG |
GTCTCATAATGTTGGCTTTGTGGACAG |
|
aco-2 |
GGCTATGCTCCAATTCATCAGC |
CCATGTTGCAGATGGTTCCCATTC |
|
idh-1 |
CTCAAAAGATCCAAGGAGGAGACATC |
CCCGGATAGAATCATCGGTATTGTAC |
|
cox-5B |
GGCTCAACTTGCTAAGACGG |
CATCTCTTTGGATCTCCTTTGCG |
|
cox-4 |
CAACTTGCTAAGACGGCTGTTG |
CACTTCTTGTTCTCGTAATCGTAGTG |
|
tba-1 |
GTTTTCAACATGCGTGAGGTCATC |
GGGGCTGGGTAGATGGAGAATTC |
2.6. Statistical analysis
All statistical evaluations were conducted in GraphPad Prism 10 (La Jolla, CA). Lifespan and pharyngeal pumping assays were compared across groups via one-way ANOVA. Data represent the mean ± standard error of the mean (SEM) from triplicate biological replicates. Significance (* P < 0.05, ** P < 0.01, *** P < 0.001).
3. Results
3.1. Metformin and rapamycin significantly extended the lifespan of C. elegans
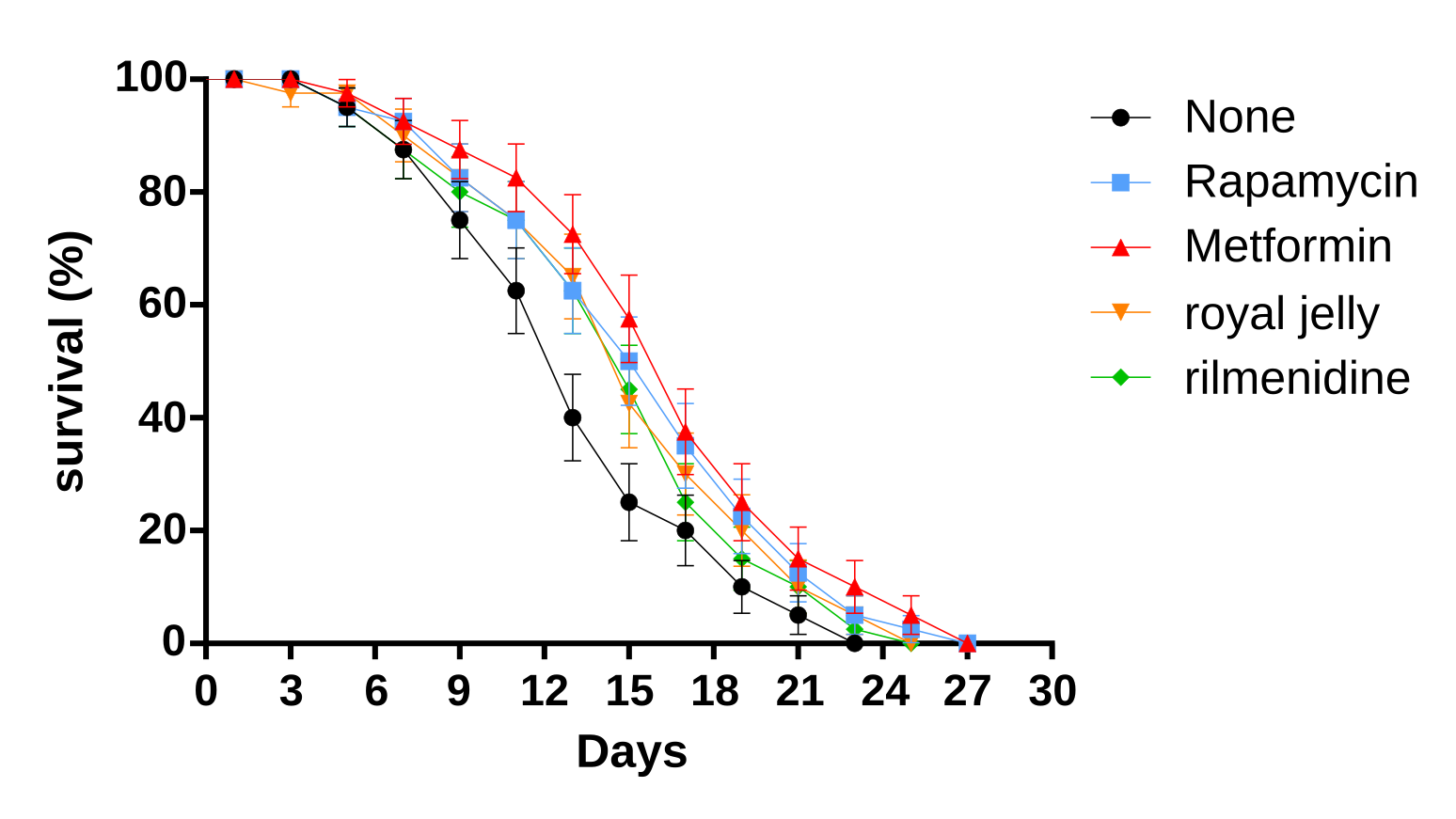
For the wild-type worms (control group), the median survival duration is 13 days. The median survival of rapamycin is 16 days, and it significantly extended lifespan 23% compared to the control group (Log Rank test, P = 0.0338, odds ratio (OR) = 1.231, 95% CI 0.7940, 1.908) (Table 3, Figure 2). Metformin significantly extended lifespan 30% more than the control group (Log Rank test, P = 0.0040, odds ratio (OR) = 1.308, 95% CI 0.8437, 2.027). Royal jelly numerically extended its lifespan by 15% more than the control group (Log Rank test, P = 0.0610, odds ratio (OR) = 1.154, 95% CI 0.7444, 1.788). Rilmenidine numerically extended lifespan by 15% more than the control group (Log Rank test, P = 0.12, odds ratio (OR) = 1.154, 95% CI 0.74, 1.79). This result suggested the strong effect of rapamycin and metformin on extending the lifespan of C. elegans.
|
Groups |
Hazard Ratio (Mantel-Haenszel) |
Hazard Ratio (log-rank) |
||
|
Hazard Ratio |
95% CI |
Hazard Ratio |
95% CI |
|
|
Rapamycin vs. control |
0.5672 |
0.3360, 0.9575 |
0.6686 |
0.4275,1.04 |
|
Metformin vs. control |
0.4585 |
0.2700,0.77 |
0.5793 |
0.3677,0.91 |
|
Royal jelly vs. control |
0.6072 |
0.3603,1.02 |
0.7008 |
0.4990,1.09 |
|
Rilmenidine vs. control |
0.6634 |
0.3936,1.11 |
0.7474 |
0.4799,1.16 |
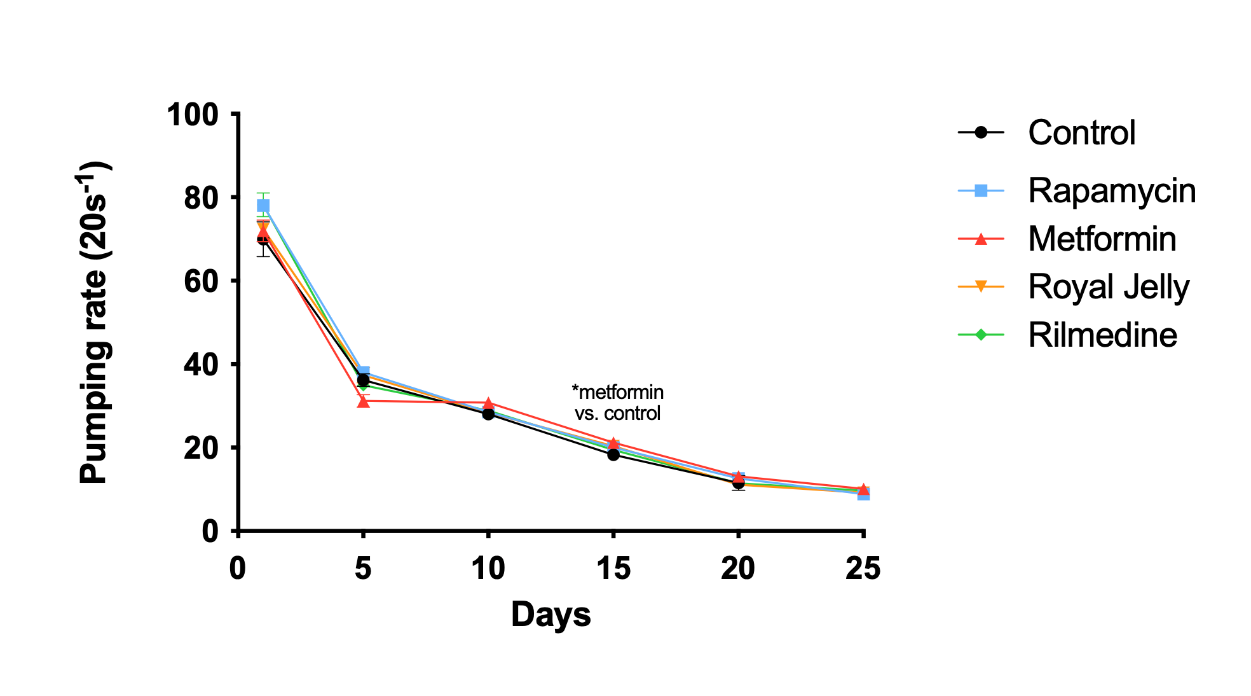
3.2. Metformin significantly enhanced the pumping rate of C. elegans
Treatment by metformin significantly increased the pumping rate at D15 compared with the control group (P = 0.0276), while rapamycin, royal jelly, and rilmenidine showed a comparable effect with the control group (Figure 3). Thus, metformin may slow down the aging process of C. elegans.
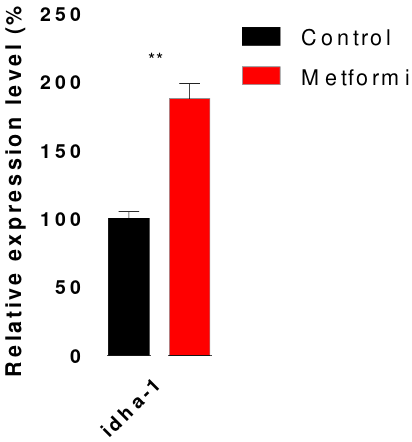
3.3. Metformin enhanced the mRNA expression level of metabolic genes
To investigate changes in the relative expression of aging-related genes after treatment with metformin, RT-qPCR was used to measure mRNA levels, with tba-1 as the inner control (Figure 4). The tested genes were aging-regulated genes and associated with metabolism, including daf-2, age-1, mecr-1, rml-1, idha-1, aco-2, idh-1, cox-5B, and cox-4 (Hamilton B, et al., A systematic RNAi screen for longevity genes in C. elegans, Genes Dev., 2005). After metformin treatment, the mRNA expression levels of idha-1 were significantly upregulated by 87% (P = 0.002).
4. Discussion
This study demonstrated that metformin and rapamycin had significant anti-aging effects in C. elegans. Rapamycin extended the lifespan by 23%, while metformin extended it by 30%, as compared to the wild-type worms. In addition, metformin enhanced the health span of C. elegans by significantly increasing pumping rates. The genetic assay uncovered its mechanism in anti-aging by upregulating metabolic genes.
4.1. The effect and mechanism of rapamycin, metformin, royal jelly, and rilmenidine on extending lifespan in C. elegans
Rapamycin, originally developed as an antifungal agent [13], has demonstrated significant lifespan-extending properties across multiple model organisms through its inhibition of the mTOR pathway. In C. elegans, genetic suppression of mTOR components (let-363/TOR or daf-15/RAPTOR) via RNAi has been shown to prolong lifespan [14], highlighting the pathway's conserved role in aging. The mTOR pathway interacts with insulin signaling to regulate development, metabolism, and longevity [15]. Its broad influence on aging-related processes - including autophagy, mitochondrial function, proteostasis, and cellular senescence [13] - makes it a key target for longevity research. However, rapamycin's potential immunosuppressive effects and hepatotoxicity raise concerns for therapeutic use [13], necessitating further investigation into its safety profile for anti-aging applications.
Metformin, a first-line antidiabetic drug with a well-established safety profile [16], has emerged as a promising geroprotective compound. It extends lifespan by 10-40% in nematodes and rodents through multiple mechanisms, such as AMPK activation, mTOR inhibition, and reduction of oxidative stress [16]. Beyond lifespan extension, metformin has been shown to improve cognitive function and mitigate neurodegeneration [16], underscoring its potential as a multifaceted anti-aging intervention. Despite its demonstrated benefits, the precise molecular pathways underlying its geroprotective effects remain unclear [16], posing a challenge for its translation into clinical longevity therapies.
Royal jelly, a natural bee product rich in bioactive compounds [17], has exhibited variable effects on lifespan across different studies. While some animal models suggest it may extend longevity through antioxidative and immunomodulatory mechanisms [17], our findings in C. elegans did not show significant anti-aging effects. This discrepancy may stem from variations in royal jelly composition, dosing, or species-specific responses. Additionally, its complex and poorly standardized formulation, along with potential estrogenic activity [17], complicates its therapeutic potential. Further research is needed to clarify its mechanisms and establish standardized preparations before it can be considered a reliable anti-aging intervention.
Rilmenidine, an imidazoline receptor agonist, showed no significant benefits in this study, although other research suggests it may extend health span in C. elegans [18]. This effect is thought to occur through activating ERK signaling pathways and inducing autophagy-related genes, which improve stress resistance and protein homeostasis [18]. Inconsistencies in results may depend on the dosage and timing of rilmenidine treatment. For therapeutic applications, rilmenidine faces limitations such as variability in receptor distribution and potential off-target effects in more complex organisms [18].
4.2. Metabolism regulates aging
Metabolism plays a key role in aging, with metabolic reprogramming driving impaired fitness, increased disease susceptibility, reduced stress response, and frailty. Tissue-specific needs, cross-organ metabolite communication, and effects on epigenetics and redox regulation influence aging-related metabolic changes, though not all are causative. Studies in model organisms link metabolic changes to lifespan and show that targeting specific pathways or altering metabolite levels can improve healthspan and extend lifespan due to the conserved nature of metabolism [19]. The manipulation of genes and metabolites in the tricarboxylic acid (TCA) cycle plays a role in regulating lifespan [20]. Whereas lifespan extension occurs when adding α-ketoglutarate, which is a metabolite generated by the TCA cycle, in C. elegans or Drosophila [21]. Isocitrate dehydrogenase (IDH) is an inhibitor enzyme that catalyzes isocitrate to become α-ketoglutarate in the TCA cycle [22]. IDH composes three isoenzymes, IDH1, IDH2, and IDH3, which are conserved in eukaryotes. IDH1 and IDH2 are NADP+ dependent, whereas IDH3 is NAD+ dependent. In humans, which are also eukaryotes, IDH3 is a heterotetramer containing two alpha subunits (IDH3A), one beat subunit (IDH3B), and one gamma subunit (IDH3G) [21]
4.3. Metformin extended the lifespan of C. elegans by enhancing the expression of idha-1
IDHA-1 is an isocitrate dehydrogenase alpha-1 in the TCA cycle in C. elegans, the ortholog of human IDH3A [23]. In this study, the mRNA expression level increased significantly after metformin treatment, along with the enhancement of lifespan in C. elegans. The previous study reported that the transgenic overexpression of idha-1 extends lifespan, increases the levels of NADPH/NADP+ratio, and elevates the tolerance to oxidative stress, which is consistent with our findings. Conversely, RNAi knockdown of idha-1 exhibits the opposite effects [21]. In the mammalian model zebrafish, metformin treatment enhanced the expression of IDH3A significantly at 390 ng/L and 2,929 ng/L in the females, but not in the males [24]. Thus, Metformin may target on idha-1 to extend lifespan, by inhibiting oxidative stress (Figure 5).
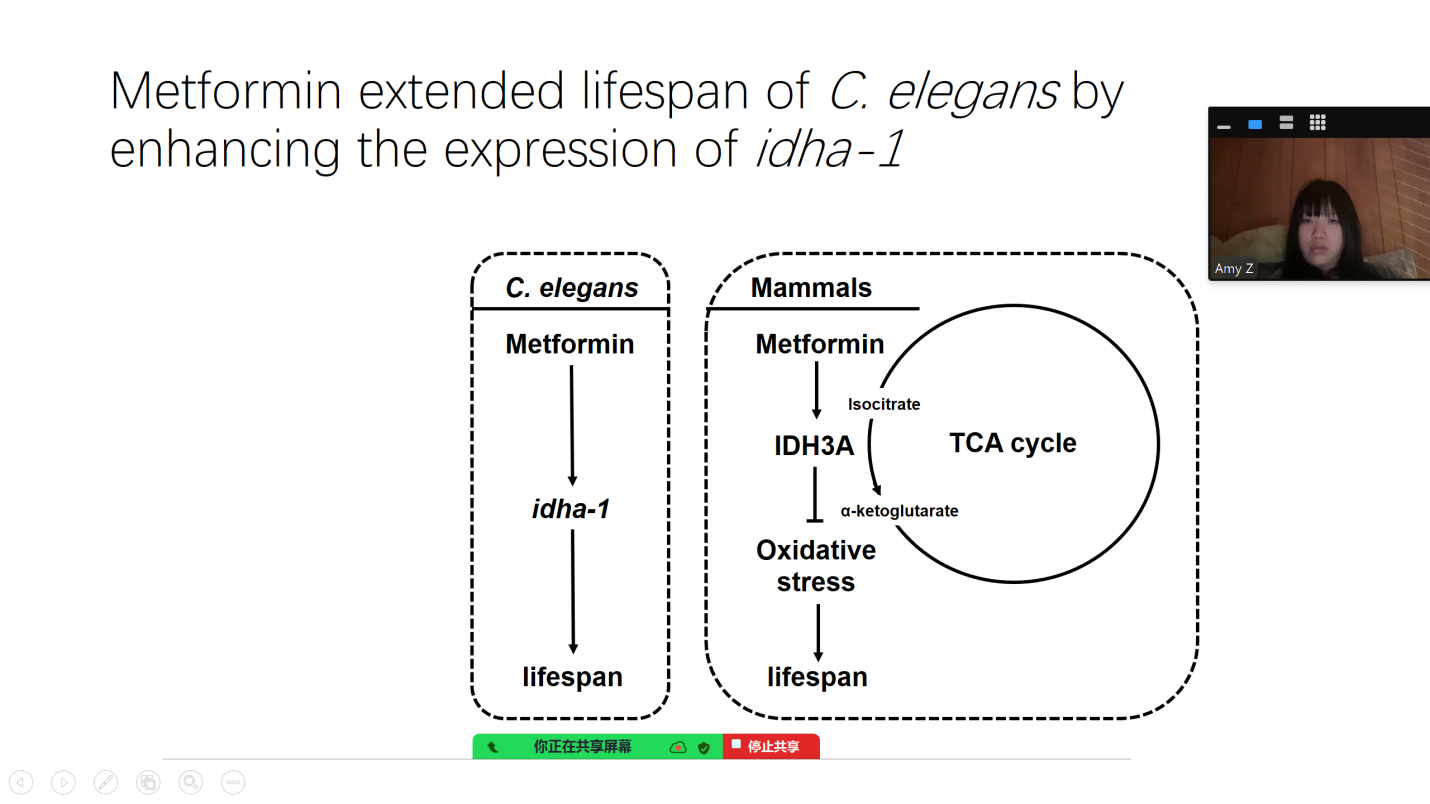
4.4. The value of this study
The current market lacks effective drugs that specifically target aging and extend healthy lifespans. This gap motivated this study to test four different drugs and compounds to explore their potential to promote longevity. Among the compounds tested, metformin, a drug commonly used to treat type 2 diabetes, was found to significantly extend lifespan. This discovery highlights its potential as a candidate for anti-aging therapy
Metformin appears to work through a specific biological pathway. It upregulates an enzyme in the TCA (tricarboxylic acid) cycle known as idha-1 [21]. This upregulation may enhance the body's tolerance to oxidative stress, a key factor in cellular aging and age-related diseases [21]. The enzyme idha-1 is identified as a promising target for future studies. It offers potential for the development of new drugs aimed at prolonging lifespan by improving cellular resilience to aging-related stressors [23]. This research has significant implications for society. By promoting a healthy lifespan, the study suggests that drugs like metformin could help prevent or delay the onset of aging-related diseases, and may help prevent aging-related diseases, such as cardiovascular diseases, neurodegenerative conditions, and cancer. Furthermore, prolonging healthy life could alleviate the social and economic burdens associated with aging populations, such as healthcare costs and caregiving demands.
5. Conclusion
The study demonstrates that metformin can extend the health span in C. elegans, potentially through the upregulation of the enzyme idha-1 in the TCA cycle. This finding lays the groundwork for future research into anti-aging drugs and offers hope for addressing age-related diseases and promoting healthy aging at both the individual and societal levels.
References
[1]. Kaeberlein, M. (2018). "How healthy is the healthspan concept?" Geroscience 40(4): 361-364.
[2]. Le Couteur, D. G. and J. Thillainadesan (2022). "What Is an Aging-Related Disease? An Epidemiological Perspective." J Gerontol A Biol Sci Med Sci 77(11): 2168-2174.
[3]. Murphy, C. T. and P. J. Hu (2013). "Insulin/insulin-like growth factor signaling in C. elegans." WormBook: 1-43.
[4]. Vagero, D., et al. (2018). "Why is parental lifespan linked to children's chances of reaching a high age? A transgenerational hypothesis." SSM Popul Health 4: 45-54.
[5]. Zhang, Y. P., et al. (2022). "Intestine-specific removal of DAF-2 nearly doubles lifespan in Caenorhabditis elegans with little fitness cost." Nat Commun 13(1): 6339.
[6]. Martel, J., et al. (2020). "Plant and fungal products that extend lifespan in Caenorhabditis elegans." Microb Cell 7(10): 255-269.
[7]. Pan, H. and T. Finkel (2017). "Key proteins and pathways that regulate lifespan." J Biol Chem 292(16): 6452-6460.
[8]. Navabpour, S., et al. (2020). "A neuroscientist's guide to transgenic mice and other genetic tools." Neurosci Biobehav Rev 108: 732-748.
[9]. Mitani, S. (2017). "Comprehensive functional genomics using Caenorhabditis elegans as a model organism." Proc Jpn Acad Ser B Phys Biol Sci 93(8): 561-577.
[10]. Brenner, S., & Gratzer, W. (1997). Loose ends. Nature, 390(6655), 35-35.
[11]. Lin, Z., et al. (2023). "Transcriptome analysis of the adenoma-carcinoma sequences identifies novel biomarkers associated with development of canine colorectal cancer." Front Vet Sci 10: 1192525.
[12]. Steven, R., et al. (2005). "The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans." Genes Dev 19(17): 2016-2029.
[13]. Papadopoli, D., et al. (2019). "mTOR as a central regulator of lifespan and aging." F1000Res 8.
[14]. Fabrizio, P., et al. (2001). "Regulation of longevity and stress resistance by Sch9 in yeast." Science 292(5515): 288-290.
[15]. Vellai, T., et al. (2003). "Genetics: influence of TOR kinase on lifespan in C. elegans." Nature 426(6967): 620.
[16]. Barzilai, N., et al. (2016). "Metformin as a Tool to Target Aging." Cell Metab 23(6): 1060-1065.
[17]. Orsolic, N. and M. Jazvinscak Jembrek (2024). "Royal Jelly: Biological Action and Health Benefits." Int J Mol Sci 25(11).
[18]. Bennett, D. F., et al. (2023). "Rilmenidine extends lifespan and healthspan in Caenorhabditis elegans via a nischarin I1-imidazoline receptor." Aging Cell 22(2): e13774.
[19]. Parkhitko, A. A., et al. (2020). "Targeting metabolic pathways for extension of lifespan and healthspan across multiple species." Ageing Res Rev 64: 101188.
[20]. Katewa, S. D., et al. (2014). "Mitobolites: the elixir of life." Cell Metab 20(1): 8-9.
[21]. Lin, Z. H., et al. (2022). "Isocitrate Dehydrogenase Alpha-1 Modulates Lifespan and Oxidative Stress Tolerance in Caenorhabditis elegans." Int J Mol Sci 24(1).
[22]. Gabriel, J. L., et al. (1986). "Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria." Metabolism 35(7): 661-667.
[23]. Kishore, R., et al. (2020). "Automated generation of gene summaries at the Alliance of Genome Resources." Database (Oxford) 2020.
[24]. Barros, S., et al. (2022). "Metformin disrupts Danio rerio metabolism at environmentally relevant concentrations: A full life-cycle study." Sci Total Environ 846: 157361.
Cite this article
Zhu,S. (2025). The Anti-Aging Effect of Metformin on C. elegans Through Idha-1, an Isocitrate Dehydrogenase in the TCA Cycle. Theoretical and Natural Science,126,21-29.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Kaeberlein, M. (2018). "How healthy is the healthspan concept?" Geroscience 40(4): 361-364.
[2]. Le Couteur, D. G. and J. Thillainadesan (2022). "What Is an Aging-Related Disease? An Epidemiological Perspective." J Gerontol A Biol Sci Med Sci 77(11): 2168-2174.
[3]. Murphy, C. T. and P. J. Hu (2013). "Insulin/insulin-like growth factor signaling in C. elegans." WormBook: 1-43.
[4]. Vagero, D., et al. (2018). "Why is parental lifespan linked to children's chances of reaching a high age? A transgenerational hypothesis." SSM Popul Health 4: 45-54.
[5]. Zhang, Y. P., et al. (2022). "Intestine-specific removal of DAF-2 nearly doubles lifespan in Caenorhabditis elegans with little fitness cost." Nat Commun 13(1): 6339.
[6]. Martel, J., et al. (2020). "Plant and fungal products that extend lifespan in Caenorhabditis elegans." Microb Cell 7(10): 255-269.
[7]. Pan, H. and T. Finkel (2017). "Key proteins and pathways that regulate lifespan." J Biol Chem 292(16): 6452-6460.
[8]. Navabpour, S., et al. (2020). "A neuroscientist's guide to transgenic mice and other genetic tools." Neurosci Biobehav Rev 108: 732-748.
[9]. Mitani, S. (2017). "Comprehensive functional genomics using Caenorhabditis elegans as a model organism." Proc Jpn Acad Ser B Phys Biol Sci 93(8): 561-577.
[10]. Brenner, S., & Gratzer, W. (1997). Loose ends. Nature, 390(6655), 35-35.
[11]. Lin, Z., et al. (2023). "Transcriptome analysis of the adenoma-carcinoma sequences identifies novel biomarkers associated with development of canine colorectal cancer." Front Vet Sci 10: 1192525.
[12]. Steven, R., et al. (2005). "The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans." Genes Dev 19(17): 2016-2029.
[13]. Papadopoli, D., et al. (2019). "mTOR as a central regulator of lifespan and aging." F1000Res 8.
[14]. Fabrizio, P., et al. (2001). "Regulation of longevity and stress resistance by Sch9 in yeast." Science 292(5515): 288-290.
[15]. Vellai, T., et al. (2003). "Genetics: influence of TOR kinase on lifespan in C. elegans." Nature 426(6967): 620.
[16]. Barzilai, N., et al. (2016). "Metformin as a Tool to Target Aging." Cell Metab 23(6): 1060-1065.
[17]. Orsolic, N. and M. Jazvinscak Jembrek (2024). "Royal Jelly: Biological Action and Health Benefits." Int J Mol Sci 25(11).
[18]. Bennett, D. F., et al. (2023). "Rilmenidine extends lifespan and healthspan in Caenorhabditis elegans via a nischarin I1-imidazoline receptor." Aging Cell 22(2): e13774.
[19]. Parkhitko, A. A., et al. (2020). "Targeting metabolic pathways for extension of lifespan and healthspan across multiple species." Ageing Res Rev 64: 101188.
[20]. Katewa, S. D., et al. (2014). "Mitobolites: the elixir of life." Cell Metab 20(1): 8-9.
[21]. Lin, Z. H., et al. (2022). "Isocitrate Dehydrogenase Alpha-1 Modulates Lifespan and Oxidative Stress Tolerance in Caenorhabditis elegans." Int J Mol Sci 24(1).
[22]. Gabriel, J. L., et al. (1986). "Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria." Metabolism 35(7): 661-667.
[23]. Kishore, R., et al. (2020). "Automated generation of gene summaries at the Alliance of Genome Resources." Database (Oxford) 2020.
[24]. Barros, S., et al. (2022). "Metformin disrupts Danio rerio metabolism at environmentally relevant concentrations: A full life-cycle study." Sci Total Environ 846: 157361.









