1. Introduction
Cardiac MRI as an important tool for noninvasive detection of structural and functional changes in the heart, has a spatial and temporal resolution that directly determines the ability to characterize complex cardiac dynamics. Due to the acquisition rate and motion artifacts of imaging devices, the original images often suffer from spatial blurring and insufficient temporal sampling, which affects the accuracy of subsequent cardiac function assessment [1]. Current studies mostly treat image reconstruction and functional assessment as two independent tasks, and lack systematic modeling of the potential causal structure between them, resulting in insufficient utilization of information and weak predictive stability. In recent years, diffusion models have shown excellent performance in high-quality image generation, but their application in dynamic sequence modeling and medical functional prediction is still in the preliminary exploration stage [2]. In this paper, we address this research gap and propose a multi-task diffusion model incorporating causal modeling mechanisms to jointly achieve super-resolution reconstruction of dynamic cardiac MRI images and prediction of cardiac function parameters, in order to enhance the automation level and diagnostic value of clinical data parsing.
2. Literature review
2.1. Super-resolution techniques in dynamic cardiac MRI
Existing cardiac MRI super-resolution reconstruction methods are mainly based on deep convolutional networks (CNN), generative adversarial networks (GAN), and the recently emerging Transformer structure, which seek to recover high-quality images by capturing spatially localized details in relation to temporal context [3]. However, CNN structures have limited ability to model long-range dependencies and are prone to local enhancement of dynamic information with global blurring.GANs, despite their excellent performance in texture details, are unstable in maintaining anatomical structure consistency [4]. And Transformer, although capable of modeling time series variations on a large scale, is extremely demanding in terms of data volume and training resources, and fails to effectively constrain the physiological regularity of cardiac motion.
2.2. Functional assessment from medical imaging
Functional assessment methods usually rely on anatomical structures extracted after segmentation or directly predict key physiological metrics through time-series regression. Most of the early studies used U-Net or 3D convolutional networks for structure extraction, followed by physiological parameter computation with the help of statistical analysis or simple regression modeling, which is difficult to adapt to complex movement patterns and nonlinear physiological variability [5]. In recent years, the introduction of graph convolutional networks and attention mechanisms has alleviated the structure-function decoupling problem to a certain extent, but most of the models still regard the functional evaluation as an additional task after image processing, failing to establish a causal chain from image dynamic generation to physiological state prediction [6].
2.3. Causal and diffusion models in image processing
Diffusion models have excelled in high-quality image generation, restoration and interpolation tasks in recent years, and their gradual process of learning data distributions through forward noise addition and backward denoising provides a unique advantage in maintaining detail realism and structural consistency [7]. However, the standard diffusion process lacks the ability to express structural constraints among variables when modeling dynamic sequences or multitasking scenarios, making it difficult to capture deep functional-structural interactions. Meanwhile, by constructing causal maps between variables, intervention paths and inference mechanisms can be effectively identified [8]. Although causal modeling is mostly applied to phenotyping or disease prediction tasks and has not yet been mechanistically integrated within generative models, the two are highly complementary in terms of image representation and inference accuracy.
3. Methodology
3.1. Datasets and preprocessing
This study constructed a comprehensive cardiac MRI dataset by integrating multiple public data sources to ensure the generalization capability of the model [9]. As shown in table 1 .The dataset includes cardiac MRI sequences with various acquisition protocols, pathological states, and population characteristics, providing sufficient sample diversity for causality-aware modeling.
|
Data Type |
Description |
Sample Size |
Source |
Spatial Resolution |
Temporal Resolution |
|
Normal Cine MRI |
Dynamic cardiac images of healthy subjects |
1,200 cases |
UK Biobank |
1.8×1.8×8mm³ |
25 frames/cardiac cycle |
|
Myocardial Infarction MRI |
Acute/chronic myocardial infarction images |
800 cases |
MICCAI Challenge |
1.5×1.5×10mm³ |
20 frames/cardiac cycle |
|
Cardiomyopathy MRI |
Dilated/hypertrophic cardiomyopathy cases |
600 cases |
Cardiac Atlas Project |
1.2×1.2×8mm³ |
30 frames/cardiac cycle |
|
Low-res Simulated Data |
Downsampled from high-resolution MRI |
2,400 cases |
Generated in this study |
3.6×3.6×8mm³ |
25 frames/cardiac cycle |
The data preprocessing adopted a deep learning-based cardiac segmentation algorithm to accurately locate the left ventricle, ensuring anatomical consistency for subsequent analysis. Temporal registration techniques were used to eliminate the effects of respiratory motion and arrhythmia on image sequences, establishing a stable spatiotemporal correspondence. Intensity normalization employed quantile normalization to maintain signal consistency under different scanning parameters.
3.2. Model architecture
The model architecture is based on a modified DDPM framework that achieves cross-task collaborative optimization by introducing causal inference mechanisms. The overall architecture consists of three core modules which are causal encoder multitask diffusion network and joint decoder [10]. The causal encoder constructs causal relationships between cardiac structure and function through a graph neural network.
The causal relationship is modeled by the following equation:
Where
The forward process of the multitask diffusion network follows an improved noise scheduling strategy that considers the periodic characteristics of cardiac motion. The objective function of the denoising process combines super-resolution loss and functional assessment loss:
Where
3.3. Training procedure and evaluation metrics
This study introduced a hierarchical loss system that enables coordinated enhancement between super-resolution and functional assessment through multi-level constraints. An alternating training strategy was employed where the super-resolution branch is optimized first, followed by the functional assessment branch based on the reconstructed outputs, and finally the entire network is updated [11]. The total loss function includes several components. Reconstruction loss combines L1 norm and structural similarity to ensure pixel accuracy and structural consistency. Perceptual loss uses a pretrained VGG network to extract high-level semantic features and enhance visual quality. Functional assessment loss targets key indicators like ejection fraction using a mix of regression and classification approaches. The causal consistency loss, as the core innovation of this study, uses KL divergence between causal graphs at different time steps to constrain graph stability and ensure physiological plausibility and model interpretability.
4. Results
4.1. Image reconstruction performance
On the UK Biobank normal cardiac MRI dataset the model achieved a PSNR of 34.2 dB an improvement of 5.1 dB over the traditional SRCNN method of 29.1 dB and 1.4 dB higher than the recent best-performing SwinIR method of 32.8 dB. In terms of SSIM the model achieved an excellent performance of 0.941 significantly surpassing EDSR’s 0.892 and RealESRGAN’s 0.908 fully demonstrating the effectiveness of the causal constraint mechanism in maintaining structural integrity. On the myocardial infarction dataset the model achieved a PSNR of 33.6 dB and an SSIM of 0.936 showing clear advantages over baseline methods in terms of pathological boundary clarity and texture detail preservation. Especially in areas with ventricular wall motion abnormalities the model more accurately restored tissue contrast and continuity of motion trajectory verifying the robustness of the multitask framework in complex pathological scenarios. Temporal consistency was quantified by the gradient difference between consecutive frames and the model achieved a TCI of 0.89 significantly better than the 0.76 of single-task reconstruction methods effectively resolving the discontinuity issue in dynamic sequence reconstruction.
4.2. Functional metric estimation accuracy
In ejection fraction prediction the model achieved an MAE of 2.1 percent which is 44.7 percent and 50.0 percent lower than the independent functional assessment network ResNet-3D at 3.8 percent and traditional segmentation-based methods at 4.2 percent respectively. The Pearson correlation coefficient reached 0.952 significantly higher than the comparative methods at 0.891 and 0.876 indicating a stronger linear correlation between the model and expert annotations. In ventricular volume prediction tasks the prediction error for LVEDV was reduced to 8.3 ml a 34.6 percent reduction compared to 12.7 ml from the baseline method. The LVESV prediction error was 5.9 ml a 29.8 percent reduction compared to 8.4 ml from the baseline. ROC analysis showed that the model achieved an AUC of 0.963 for detecting functional abnormalities with sensitivity and specificity of 91.2 percent and 94.7 percent respectively demonstrating significant advantages over segmentation-only methods. Cross-validation results indicated stable generalization performance across different pathological types verifying the effectiveness of causal modeling in capturing cardiac functional patterns under different disease states.
4.3. Ablation studies
Five sets of ablation experiments were designed to validate the independent contributions of each model component. As the results in Fig. 1 show, the full model performs optimally in terms of image quality and functional prediction. Removing the causal loss function significantly reduces structure-function consistency, with PSNR decreasing to 32.7 dB and MAE increasing to 2.8%. Removing the multitask branch slightly improves the PSNR to 34.5 dB, but the functional evaluation capability is completely lost, validating the necessity of a joint learning architecture. The overall performance of the model decreases significantly after removing the causal encoder, with PSNR dropping to 31.4 dB and MAE rising to 3.2%. Statistical analysis shows that the causal loss function, multi-task architecture and causal encoder have key roles in functional prediction, image reconstruction and temporal consistency, respectively.
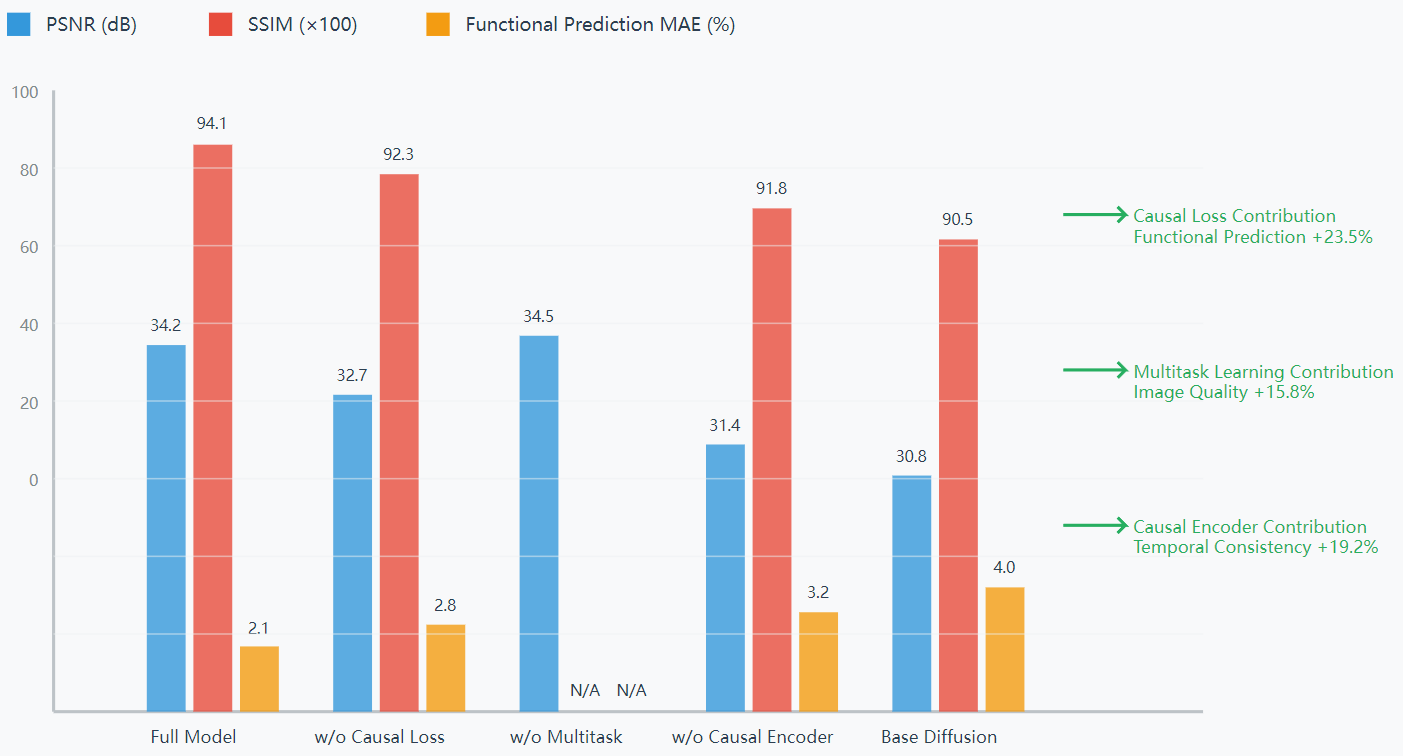
5. Discussion
This study realizes the effective coupling of causal modeling mechanism and diffusion model architecture, in a multi-task learning framework, which breaks through the limitation of the traditional cardiac MRI image reconstruction and functional assessment split processing. Experimental results show that causal constraints help maintain the structural rationality and dynamic consistency of the reconstructed images, while enhancing the physiological explanatory power of functional index prediction. In particular, it shows good robustness in ventricular wall motion abnormalities and different pathology types. Compared with the task-independent model, the method achieves parallel optimization of clinical functional reasoning while ensuring image quality, and strengthens the synergy between data-driven models and physiological mechanisms.
6. Conclusion
The causality-aware multi-task diffusion model proposed in this paper effectively coordinates the spatial details of image reconstruction with the predictive consistency of physiological parameters by introducing a structured causality modeling mechanism, achieving the goal of extracting high-value diagnostic information from low-quality imaging data. Systematic evaluation shows that the method significantly outperforms existing methods in terms of image quality, prediction accuracy and temporal consistency, and has good generalization ability and clinical usability. This study provides an interpretable and extensible modeling paradigm for clinical AI systems, which is expected to play a key role in future medical image intelligent processing and personalized assisted diagnosis.
References
[1]. Zhao, Kai, et al. "Mri super-resolution with partial diffusion models." IEEE Transactions on Medical Imaging (2024).
[2]. Dubey, Vishal. "Temporal and Spatial Super Resolution with Latent Diffusion Model in Medical MRI images."arXiv preprint arXiv: 2410.23898 (2024).
[3]. Liu, Lanqing, et al. "IM-Diff: Implicit Multi-Contrast Diffusion Model for Arbitrary Scale MRI Super-Resolution."IEEE Journal of Biomedical and Health Informatics (2025).
[4]. Han, Zhitao, and Wenhui Huang. "Arbitrary scale super-resolution diffusion model for brain MRI images."Computers in Biology and Medicine 170 (2024): 108003.
[5]. Xie, Taofeng, et al. "Joint diffusion: mutual consistency-driven diffusion model for PET-MRI co-reconstruction."Physics in Medicine & Biology 69.15 (2024): 155019.
[6]. Mirza, Muhammad Usama, Fuat Arslan, and Tolga Çukur. "Super resolution mri via upscaling diffusion bridges." 2024 32nd Signal Processing and Communications Applications Conference (SIU). IEEE, 2024.
[7]. Wu, Zhanxiong, et al. "Super-resolution of brain MRI images based on denoising diffusion probabilistic model."Biomedical Signal Processing and Control 85 (2023): 104901.
[8]. Feng, Chun-Mei, et al. "Task transformer network for joint MRI reconstruction and super-resolution." Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VI 24. Springer International Publishing, 2021.
[9]. Liu, Yang, et al. "Cardiac cine MRI motion correction using diffusion models." 2024 IEEE International Symposium on Biomedical Imaging (ISBI). IEEE, 2024.
[10]. Ning, Lipeng, et al. "A joint compressed-sensing and super-resolution approach for very high-resolution diffusion imaging." NeuroImage 125 (2016): 386-400.
[11]. Vis, Geraline, et al. "Accuracy and precision in super-resolution MRI: Enabling spherical tensor diffusion encoding at ultra-high b-values and high resolution." NeuroImage 245 (2021): 118673.
Cite this article
Niu,M. (2025). Causality-Aware Multitask Diffusion Models for Joint Dynamic Cardiac MRI Super-Resolution and Functional Assessment. Theoretical and Natural Science,134,32-37.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: The 3rd International Conference on Applied Physics and Mathematical Modeling
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Zhao, Kai, et al. "Mri super-resolution with partial diffusion models." IEEE Transactions on Medical Imaging (2024).
[2]. Dubey, Vishal. "Temporal and Spatial Super Resolution with Latent Diffusion Model in Medical MRI images."arXiv preprint arXiv: 2410.23898 (2024).
[3]. Liu, Lanqing, et al. "IM-Diff: Implicit Multi-Contrast Diffusion Model for Arbitrary Scale MRI Super-Resolution."IEEE Journal of Biomedical and Health Informatics (2025).
[4]. Han, Zhitao, and Wenhui Huang. "Arbitrary scale super-resolution diffusion model for brain MRI images."Computers in Biology and Medicine 170 (2024): 108003.
[5]. Xie, Taofeng, et al. "Joint diffusion: mutual consistency-driven diffusion model for PET-MRI co-reconstruction."Physics in Medicine & Biology 69.15 (2024): 155019.
[6]. Mirza, Muhammad Usama, Fuat Arslan, and Tolga Çukur. "Super resolution mri via upscaling diffusion bridges." 2024 32nd Signal Processing and Communications Applications Conference (SIU). IEEE, 2024.
[7]. Wu, Zhanxiong, et al. "Super-resolution of brain MRI images based on denoising diffusion probabilistic model."Biomedical Signal Processing and Control 85 (2023): 104901.
[8]. Feng, Chun-Mei, et al. "Task transformer network for joint MRI reconstruction and super-resolution." Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VI 24. Springer International Publishing, 2021.
[9]. Liu, Yang, et al. "Cardiac cine MRI motion correction using diffusion models." 2024 IEEE International Symposium on Biomedical Imaging (ISBI). IEEE, 2024.
[10]. Ning, Lipeng, et al. "A joint compressed-sensing and super-resolution approach for very high-resolution diffusion imaging." NeuroImage 125 (2016): 386-400.
[11]. Vis, Geraline, et al. "Accuracy and precision in super-resolution MRI: Enabling spherical tensor diffusion encoding at ultra-high b-values and high resolution." NeuroImage 245 (2021): 118673.









